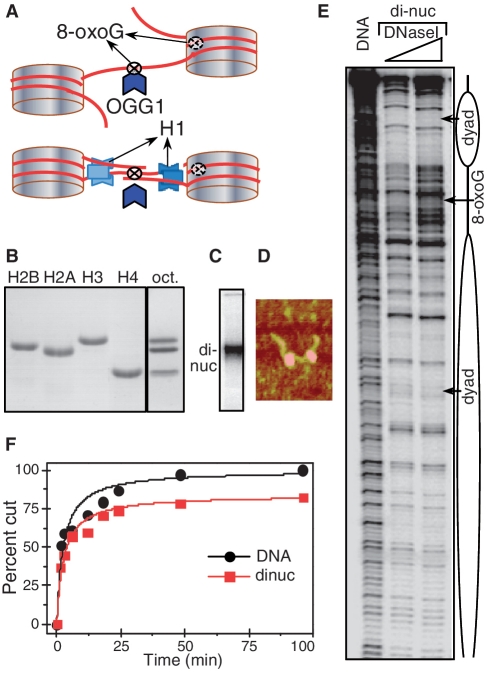
Figure 1.
The 8-oxoG located in the linker DNA of the dinucleosome is removed by OGG1 as efficiently as in free DNA. (A) Schematics of the 601 dinucleosome constructs used for studying the repair of 8-oxoG. Tandem repeats of two 601 DNA sequences were used to reconstitute positioned dinucleosomes with linker DNA (between the two nucleosomes) of length of either 20 or 75 bp. The flanking free DNA arms at both ends of the dinucleosome were of 52 and 56 bp, respectively (for details see Supplementary Figure S1). 8-oxoG was inserted either in close vicinity to the nucleosomal dyad or in the centre of the linker DNA. Experiments were carried out either in the absence or in the presence of histone H1. (B) Eighteen percent SDS gel electrophoresis of the purified recombinant histones (left); right (oct), the equimolar mixture of the core histones used for the reconstitution of the dinucleosome. (C–E): Characterization of the reconstituted dinucleosome by EMSA (C), AFM (D) and DNase I footprinting (E). (F): Time course of the digestion of free DNA and dinucleosomes. Equal amounts of 32P-end labelled free DNA and dinucleosomes were incubated with 0.5 U of OGG1 for the times as indicated. The reaction was stopped and DNA was isolated by phenol–chloroform treatment. The products of the OGG1 cleavage reaction were separated on 8% denaturing PAGE and after exposure of the dried gel, data were quantified and expressed as a percentage of cut fractions versus incubation time. Solid curves are least squared fits of experimental points by the formula: R = (Rmt/tc)/(1 + t/tc). Rm values are 100% and 84 ± 4% for DNA and dinucleosomes respectively and tcDNA/tcdinuc = 0.94 ± 19%.