Abstract
Zinc-finger nucleases and TALE nucleases are produced by combining a specific DNA-binding module and a non-specific DNA-cleavage module, resulting in nucleases able to cleave DNA at a unique sequence. Here a new approach for creating highly specific nucleases was pursued by fusing a catalytically inactive variant of the homing endonuclease I-SceI, as DNA binding-module, to the type IIP restriction enzyme PvuII, as cleavage module. The fusion enzymes were designed to recognize a composite site comprising the recognition site of PvuII flanked by the recognition site of I-SceI. In order to reduce activity on PvuII sites lacking the flanking I-SceI sites, the enzymes were optimized so that the binding of I-SceI to its sites positions PvuII for cleavage of the composite site. This was achieved by optimization of the linker and by introducing amino acid substitutions in PvuII which decrease its activity or disturb its dimer interface. The most specific variant showed a more than 1000-fold preference for the addressed composite site over an unaddressed PvuII site. These results indicate that using a specific restriction enzyme, such as PvuII, as cleavage module, offers an alternative to the otherwise often used catalytic domain of FokI, which by itself does not contribute to the specificity of the engineered nuclease.
INTRODUCTION
Gene targeting allows the introduction of genetic elements into chosen genomic loci. The usual approach to achieve this goal is based on homologous recombination, the natural efficiency of which is quite low but can be stimulated by double-strand breaks using engineered nucleases. For gene targeting these endonucleases should recognize unique sequences and cleave DNA highly specifically, and thereby initiate double-strand break repair by induced homologous recombination with a DNA template supplied in trans (1,2).
Such nucleases with very high specificity have been engineered by rational and combinatorial strategies (3): among them are (i) zinc-finger nucleases (4–9), (ii) engineered homing endonucleases (10–13) and (iii) triple helix forming oligonucleotides linked to restriction enzymes (14) or other DNA cleavage moduls (15,16).
Combining a cleavage module with a recognition module in order to generate nucleases with high specificity was first achieved with the non-specific DNA cleavage domain of the restriction endonuclease FokI and the Ubx homeodomain (17), which was soon followed by the fusion of zinc finger motifs to the FokI cleavage domain (18). Most chimeric nucleases produced so far use the FokI cleavage domain as cleavage module (19), and the majority of them are zinc-finger nucleases. A noteworthy exception is the fusion product consisting of a catalytically inactive homing endonuclease and the FokI cleavage domain (20), which resulted in a fusion protein recognizing the homing endonuclease recognition site and cleaving the DNA in both strands 2 and 6 nt downstream from the recognition site, as expected for the Type IIS restriction endonucleases FokI. With a similar approach the 5mCpG-binding domain of a 5mCpG-specific DNA glycosylase was fused to the FokI cleavage domain, in order to obtain a chimeric nuclease that cleaves DNA at 5mCpG-sites (21). Similarly, the control protein C.BcII that represses the expression of the methyltransferase of the BcII R-M system was fused to the non-specific cleavage domain of BmrI; the chimeric nuclease recognizes specific sites in the vicinity of the C.BcII control sequence (22).
Recently, so called transcription activator like effector (TALE) nucleases (TALENs) were produced; they consist of TALEs as recognition module (23–26) and the FokI cleavage domain as cleavage module. Similarly as zinc-fingers, TALEs can be designed to recognize different DNA sequences (23,24) and therefore promise to be useful for the production of engineered nucleases of extended specificity (27–33).
To our knowledge no attempts have been made so far to produce chimeric nucleases consisting of a homing endonuclease (as a DNA-binding module) and a restriction enzyme (as a DNA binding and cleavage module, respectively). Such chimeric nucleases ideally should have an extended specificity and should recognize a bipartite or tripartite (in case of homodimeric restriction endonucleases, see Figure 1A) recognition site, comprising that of the homing endonuclease and that of the restriction endonuclease and cleave the DNA within the recognition site of the restriction endonuclease.
Figure 1.
(A) Cartoon representation of the fusion enzyme. The C-terminus of one subunit of a dimeric restriction enzyme (PvuII; blue) is fused via a peptide linker (gray) to the N-terminus of a catalytically inactive homing endonuclease (I-SceI; green). This fusion enzyme binds as a homodimer to a tripartite recognition site consisting of the PvuII recognition site (orange) flanked by two I-SceI sites (red) separated by an un-specified DNA (gray). The very C-terminal Strep-tag used for affinity purification is depicted in yellow. (B) Model of the engineered fusion enzyme. One subunit of the fusion enzyme consists of one I-SceI [green; pdb 1r7 m (40)] and one subunit of the PvuII dimer [blue, pdb 1pvi (47)]. This model was built by aligning the recognition sites from the crystal structures of the individual proteins on a DNA composed of the PvuII recognition site (orange) and two I-SceI sites (red) separated by 6 bp up- and downstream. The C-terminus of PvuII and the N-terminus of I-SceI, separated by 2.6 nm, are indicated by red spheres; this distance must be covered by a peptide linker of suitable length.
Type II restriction endonucleases are highly specific enzymes; they recognize DNA sequences of 4–8 bp in length, which means that their recognition sites will occur on average every 44–48 bp. PvuII (34), a homodimeric Type IIP restriction endonuclease, for example, recognizes the palindromic double-stranded DNA sequence CAG↓CTG and cleaves it in the presence of Mg2+ as indicated, generating blunt ends. Homing endonucleases are even more specific, as they recognize much longer sequences than restriction endonucleases (35). I-SceI (36,37), a monomeric homing endonuclease of the LAGLIDADG family, for example, recognizes an 18 bp sequence (TAGGGATAACAGGGTAAT) and in the presence of Mg2+ cleaves the DNA within this sequence. Typically, homing endonucleases cleave complex genomes at a few sites only.
A fusion protein consisting of PvuII as cleavage module and a catalytically inactive I-SceI variant (= S*) as binding module, separated by a linker Lx, for example PLxS*, which upon dimerization (viz. S*LxP/PLxS*) could be expected to recognize a 42 bp sequence. This composite site consists of one I-SceI site, one PvuII site and one reverse complement of the I-SceI site: TAGGGATAACAGGGTAAT — CAG↓CTG— ATTACCCTGTTATCCCTA. Cleavage should occur in the addressed PvuII site at the position indicated. This requires that the catalytic module, i.e. the PvuII part of the fusion protein is only activated after the DNA binding modules, the two I-SceI*parts of the fusion protein, have positioned the catalytic module vis-à-vis the scissile phosphodiester bonds of the PvuII site. The PLxS* fusion protein should not be able to cleave an unaddressed PvuII site, i.e. a PvuII site not flanked by I-SceI sites. We demonstrate here that by an appropriate choice of the linker between PvuII and I-SceI* and by introducing amino acid substitutions into PvuII and I-SceI*, the PLxS* fusion protein cleaves with high preference addressed PvuII sites.
MATERIALS AND METHODS
Construction of the PvuII-I-SceI fusions
The PvuII-I-SceI fusion enzyme and variants thereof were created by fusing scPvuII (38) or wt PvuII (39) via their C-terminus to the N-terminus of a catalytically inactive variant of I-SceI (I-SceI*) which had been truncated at the C-terminus (ΔC9). These last nine amino acid residues were not resolved in the co-crystal structure of I-SceI (40). For this construction, the genes coding for scPvuII or wt PvuII with the coding sequence for a C-terminal His6-tag were connected to the gene coding for I-SceI(D44S) (41) and introduced into the vector pASK-IBA63b-plus (IBA) coding for a C-terminal Strep-tag. For further improvement of the fusion protein two active site residues of I-SceI were exchanged (D44N and D145A) according to (20) via PCR-based site-directed mutagenesis (42). The mutagenesis of selected residues of PvuII was performed in the same way for the fusion protein and for PvuII alone (for control reasons), whose gene was also introduced into pASK-IBA63b-plus. Between the genes for PvuII and I-SceI three restriction enzyme sites (NheI, BsiWI, AgeI) were introduced, replacing the coding sequence for the His6-tag (LH → L6, where L denotes the linker). By cleaving the resulting vector with NheI and AgeI, and filling in, the linkers LN, L+ and L - were obtained. The correctness of all genetic constructs was confirmed by sequencing over the entire coding region for PvuII-Lx-I-SceI, containing the different linkers Lx (LH, L6, LN, L+ and L−).
Purification of the enzymes
The pASK-IBA63b-plus plasmids harboring the coding sequences for the fusion enzymes or PvuII alone were introduced into Escherichia coli strain XL-Gold, which had been transformed before with a plasmid encoding the PvuII methyltransferase gene (pLGM). The cells were grown at 37°C to an OD600 of 1. Protein expression was induced by adding 200 µg/l anhydrotetracycline, followed by further growth at 20°C overnight. After harvesting the cells by centrifugation the cell pellet was dissolved in 100 mM Tris–HCl, pH 8, 1 M NaCl, 1 mM EDTA, lysed by sonification and centrifuged for 30 min (>17 000 g) to remove cell debris. The recombinant Strep-tagged proteins were purified using Strep-Tactin Sepharose (IBA). After washing with a high-salt (100 mM Tris–HCl, pH 8, 1 M NaCl, 1 mM EDTA) and a low-salt (100 mM Tris–HCl, pH 8, 300 mM NaCl, 1 mM EDTA) washing buffer, the tagged proteins were eluted with 100 mM Tris–HCl, pH 8.0, 300 mM NaCl, 2.5 mM desthiobiotin. The purity of the eluates was analyzed by SDS-PAGE and selected fractions were pooled and dialysed overnight (50 mM Na-phosphate, pH 7.5, 100 mM NaCl, 0.5 mM EDTA, 60% glycerol). Protein concentration was determined by A280 measurements using a molar extinction coefficient of 92 250 M−1 cm−1 at 280 nm as calculated from the amino acid composition (43). Protein concentrations are given as dimers throughout the text.
DNA cleavage assays
DNA cleavage activity assays were carried out with 50 nM of a 454 bp PCR fragment containing one I-SceI site. This substrate was incubated with either 5U I-SceI (Fermentas) in Buffer Tango (Fermentas), 100 nM PvuII or 100 nM scPLHS* in Buffer Green (Fermentas) for 45 min at 37°C. Cleavage products were separated by 6% PAGE, stained with ethidium bromide and visualized by a BioDocAnalyze system (Biometra). The cleavage products obtained with scPLHS* were gel purified and sequenced.
For plasmid cleavage assays, the different target sequences (Table 2) were introduced into pAT153. The plasmid substrate was used in 8 nM concentration and incubated with 8 nM of the corresponding enzyme either for 3 h or overnight at 37°C. DNA cleavage by PvuII was performed in Buffer Green (Fermentas), by the fusion enzymes in an optimized potassium glutamate buffer [KGB (44)] (100 mM K-glutamate, 25 mM Tris–acetate, pH 7.5, 100 mM KCl, 0.8 mM Mg-acetate, 10 µg/ml BSA, 0.5 mM 2-mercaptoethanol). The cleavage products were separated on 0.8% agarose gels, stained with ethidium bromide and visualized by the BioDocAnalyze system. Quantification of linear product formation was performed by densitometry.
Table 2.
Overview of the substrates used
| Name | Recognition sites |
|---|---|
| S | I-SceI: TAATGGGACAATAGGGAT |
| P | PvuII: CAGCTG |
| S4P4S | I-SceI-4 bp-PvuII-4 bp-I-SceI |
| S6x6S | I-SceI-6 bp-KpnI-6 bp-I-SceI |
| S6P6S | I-SceI-6 bp-PvuII-6 bp-I-SceI |
| S6P6S_P | I-SceI-6 bp-PvuII-6 bp-I-SceI-763 bp-PvuII |
| S8P8S | I-SceI-8 bp-PvuII-8 bp-I-SceI |
The substrates P and S6P6S are also called unaddressed and addressed substrates, respectively.
For bacteriophage λ-DNA (which contains 15 PvuII sites) cleavage, the DNA concentration was chosen such, that 8 nM of PvuII sites (corresponding to 564 pM λ-DNA) were incubated with 8 nM fusion enzymes or PvuII in optimized KGB or Buffer Green, respectively, for 3 h at 37°C.
Analytical ultracentrifugation
Protein PL6S* was purified as described above followed by size exclusion chromatography on a Superdex 200 10/300 GL column with 50 mM Na-phosphate, pH 7.5, 500 mM NaCl, 0.5 mM EDTA. Fractions containing pure enzyme were pooled and one-half was dialyzed against 50 mM Na-phosphate, pH 7.5, 500 mM NaCl, 0.47 M glycerol (high salt conditions), the other half against 50 mM Na-phosphate, pH 7.5, 100 mM NaCl, 0.47 M glycerol (low salt conditions).
Sedimentation velocity runs were performed at 5°C and 45 000 r.p.m. in a Beckman Coulter Optima XL-I using the absorbance optics of the analytical ultracentrifuge. The experiments were carried out in an An-50 Ti rotor using 12 mm standard double sector centerpieces filled with 400 µl sample. Under high-salt conditions protein concentrations between 0.2 and 3.25 µM were examined, under low salt conditions a concentration of 1.75 µM was used. Depending on protein concentration, UV-absorption was measured at 230 or 280 nm, respectively. Sedimentation profiles were analyzed using the program SEDFIT (45), providing a model of diffusion-corrected differential sedimentation coefficient distributions [c(s) distributions]. Partial specific volumes, buffer densities and viscosities were determined with the program SEDNTERP (46) and used to correct the sedimentation coefficients to s20,w.
Competition cleavage experiments
To test the specificity of the fusion enzymes for the addressed site, radioactively labeled PCR-fragments using [α32P] dATP containing either the addressed site (S6P6S, for definition see Table 2) or the unaddressed site (P) were produced. 10 nM of each of these fragments were incubated together with 10 nM of the fusion enzymes in KGB (100 mM K-glutamate, 25 mM Tris–acetate, pH 7.5, 100 mM KCl, 0.8 mM Mg-acetate, 10 µg/ml BSA, 0.5 mM 2-mercaptoethanol) at 37°C; after defined time intervals samples were taken and subjected to polyacrylamide gel electrophoresis. Cleavage products were analyzed with the Instant Imager system (Packard) and quantified with the Instant Imager software. Initial rates were calculated by linear regression analysis.
Electrophoretic mobility shift assay
The affinity of the fusion enzymes to their target sequences was determined with electrophoretic mobility shift assays (EMSAs), using radioactively labeled DNA-fragments which were produced via PCR using [α32P] dATP. One fragment contained the addressed site (S6P6S) and another one only an I-SceI site (S). For KD determination 2 nM of the radioactively labeled addressed substrate were incubated with fusion enzyme in concentrations ranging from 1–150 nM in KGB without Mg2+ for 30 min at room temperature. For the unaddressed substrate the enzyme concentration ranged from 20 to 180 nM. After adding 1 µl 87% glycerol the samples were loaded onto a 6% polyacrylamide Tris–acetate, pH 8.5 gel and subjected to electrophoresis for 2 h at 10 V/cm. The bands were visualized using the Instant Imager system (Packard). The intensities of bands representing free and bound DNA were quantified with the Instant Imager software and the percentage of bound DNA was calculated. The data were fit and the KD determined using a non-linear regression analysis.
RESULTS
Design of the fusion enzyme
For creating a highly specific nuclease with extended specificity the type IIP restriction endonuclease PvuII was fused as the cleavage module to a catalytically inactive variant of the homing endonuclease I-SceI (= S*) as an additional binding module (Figure 1A). Due to the C2 symmetry of the homodimeric PvuII (= P) the fusion would result in a protein with the PvuII homodimer flanked by two I-SceI* molecules. The fusion protein, PLxS*, after homodimerization, should recognize a sequence consisting of the PvuII recognition sequence flanked by two I-SceI recognition sequences, S…P…S (where S and P denote the I-SceI and PvuII recognition sequences, respectively).
In the co-crystal structure, the N-terminal part of I-SceI is an α-helix sticking out of the otherwise compact structure of this enzyme and could serve as an attachment point for PvuII, as shown in a model of the fusion protein in complex with DNA, which was generated using the pdb coordinates for I-SceI [pdb:1r7 m (40)] and PvuII [pdb: 1pvi (47)] and ideal B-DNA (Figure 1B). In this model the target sites for I-SceI and PvuII are separated by 6 bp, which leads to all three proteins in the fusion protein approaching the DNA from the same side.
Engineering of PvuII-I-SceI fusion enzymes and finding a target with an optimum distance between the I-SceI and the PvuII recognition site
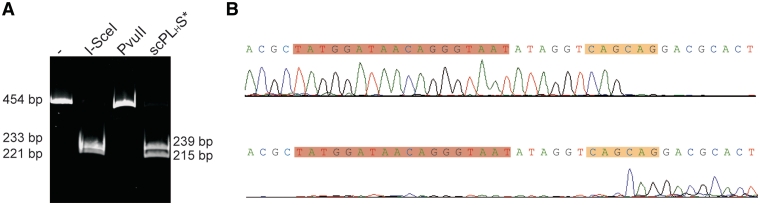
In our first construct, produced for the purpose of testing our approach, the single chain variant of PvuII, scPvuII, was fused via its C-terminal His6-tag as a linker (LH) to the catalytically inactive I-SceI D44S variant (41). The DNA cleavage activity of the scPvuII-LH-I-SceID44S fusion protein was tested on an oligonucleotide substrate containing only an I-SceI site and no PvuII site, which we expected not to be cleaved. However, a specific cleavage pattern was obtained (Figure 2A), which after sequencing of the product (Figure 2B) turned out to be due to cleavage at a PvuII ‘star’ site, (CAG↓CAG), 6 bp downstream from the I-SceI site. ‘Star’ sites, which differ in one base pair from the canonical recognition site, are only attacked by type II restriction enzymes at high concentrations of organic solvents such as DMSO and glycerol, at alkaline pH or in the presence of Mn2+. The DNA cleavage result that we obtained, therefore, showed that (i) I-SceI activates PvuII to cleave a site that it normally would not attack and that it directs PvuII to this site, and (ii) that a distance of 6 bp between the I-SceI and the PvuII recognition site in the substrate as predicted by the structural model (Figure 1B) is acceptable for the scPvuII-I-SceI D44S fusion protein. The DNA cleavage activity of this fusion protein was tested under various buffer conditions, among them the KGB (100 mM K-glutamate, 25 mM Tris–acetate, pH 7.5, 10 mM Mg-acetate, 10 µg/ml BSA, 0.5 mM 2-mercaptoethanol) which had been developed to mimic physiological conditions (44) and to which we added 100 mM KCl and in which the concentration of Mg-acetate was decreased to 0.8 mM, in order to more closely approach the KCl and Mg2+ concentration prevailing in vivo. In this buffer the largest difference in activity between scPvuII and scPLH S* (Supplementary Figure S1) was observed, which means that the fusion construct cleaves an addressed site much better than scPvuII alone does. All further cleavage experiments of the fusion enzymes were performed in the optimized version of KGB (100 mM K-glutamate, 25 mM Tris–acetate, pH 7.5, 100 mM KCl, 0.8 mM Mg-acetate, 10 µg/ml BSA, 0.5 mM 2-mercaptoethanol).
Figure 2.
(A) Cleavage of 50 nM of a 454 bp PCR fragment containing an I-SceI recognition site. Lane 1 (-): uncleaved DNA fragment; Lane 2: cleavage products obtained by incubation of the 454 bp PCR fragment with 5 U I-SceI (Fermentas) in Buffer Tango; Lanes 3 and 4: cleavage products obtained by incubation of the 454 bp PCR fragment with 100 nM PvuII and 100 nM scPLHS, respectively, in Buffer Green. (B) Sequencing results of the cleavage product of scPLHS* from (A). The fragments were gelpurified and sequenced in forward (upper panel) and reverse (lower panel) direction. The I-SceI recognition sequence is indicated by a red bar and the cleavage site, which corresponds to a PvuII ‘star’ site by an orange bar. These two sites are separated by 6 bp.
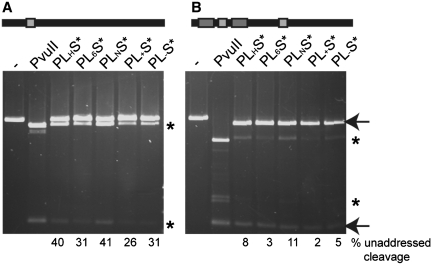
For all other constructs the homodimer of PvuII was used instead of the single chain variant, which means that the fusion proteins contains two I-SceI* binding modules rather than one as in the scPvuII fusion proteins. The two I-SceI* modules are linked via their N-termini to the C-termini of the two PvuII subunits. For this construct, we also used another I-SceI variant, namely I-SceIΔC9,D44N,D145A; this variant has amino acid substitutions in both catalytic centers and is C-terminally truncated. For the amino acid substitutions in the catalytic center it was shown before, that this combination improves the solubility of I-SceI (20). Since fusion proteins with wtPvuII are homodimeric, their recognition site will be longer and they are likely to be more specific. wtPvuII and the I-SceIΔC9,D44N,D145A were fused using different linkers (LH, L6, LN, L+ and L−) (Table 1) which all could cover the ∼26 Å distance between the C-terminus of PvuII and the N-terminus of I-SceI in our structural model (Figure 1B). These fusion proteins were expressed and purified. To test these enzymes, substrates with different distances between the I-SceI and the PvuII recognition sites (Table 2) were produced. Considering the structural model and the DNA cleavage results with scPL(H)S* cleaving 6 bp downstream from the I-SceI site, we tested the different linker variants for specificity on the linearized plasmid substrate S6P6S_P, i.e. a substrate containing a PvuII site (P) flanked by two I-SceI sites (S) in a 6 bp distance (6P and P6, respectively) and an additional single PvuII site (_P) (Figure 3B). wtPvuII cleaves this substrate at both PvuII sites (the addressed one, which is flanked by I-SceI sites, and the other one, which is unaddressed) resulting in three major products, which shows, that PvuII by itself has no preference for one of the sites due to sequence context. The fusion enzymes in contrast cleave preferentially the addressed site (S6P6S) resulting in the fragments indicated with an arrow in Figure 3B. The unaddressed site (_P) is also cleaved but much more slowly; PL6S* and PL+S* cleaved the unaddressed site with an at least two orders of magnitude slower rate than the addressed site (Table 4). Still, a plasmid substrate with only an unaddressed PvuII site (Figure 3A) which is quickly cleaved by wtPvuII is also cleaved by the fusion enzymes, albeit with a lower rate. PL6S* and PL+S* show the slowest unaddressed cleavage rate. For this reason these variants were used for further optimization and characterization.
Table 1.
Overview of the linkers used in the engineered PvuII-I-SceI fusions
| Linker name | Length | Sequence | Source |
|---|---|---|---|
| LH | 10 AA | GSHHHHHHGT | Engineered |
| L6 | 6 AA | ASRTTG | Engineered |
| LN | 10 AA | ASGGSGSGSG | Engineered |
| L+ | 10 AA | ASTKQLVKSG | FokI |
| L− | 10 AA | ASGDSGSDSG | Engineered |
Figure 3.
Cleavage activity of fusion enzymes with different linkers on linearized DNA. The DNA substrate contains either (A) a single PvuII site (light gray bar) or (B) the tripartite recognition site consisting of a PvuII site (light gray bar) flanked by two I-SceI sites (dark gray bars) each 6 bp away, and an additional unaddressed PvuII site. The experiment was performed under optimized conditions for the corresponding enzymes, namely Buffer Green for PvuII and optimized KGB for the fusion enzymes. The products of addressed cleavage at the tripartite recognition site are indicated by arrows and the products of unaddressed cleavage at PvuII sites by asterisks. The percentage of unaddressed cleavage is shown at the bottom.
Table 4.
Summary of activities on addressed and unaddressed substrates and resulting specificities determined by competition cleavage experimentsa for all tested fusion enzyme variants
| Enzyme | Activity (nM min−1) |
Specificity | |
|---|---|---|---|
| Addressed | Unaddressed | (Addressed/ unaddressed) | |
| PL6S* | 0.88 ± 0.07 | 0.0026 ± 0.0004 | 338 |
| PL+S* | 1.00 ± 0.08 | 0.014 ± 0.004 | 72 |
| P(T46G, Y94F)L6S* | 0.018 ± 0.005 | 0.0010 ± 0.0004 | 18 |
| P(T46G, Y94F)L+S* | 0.025 ± 0.009 | 0.0023 ± 0.0008 | 11 |
| P(L12E)L6S* | 0.39 ± 0.05 | <0.0001b | >1000 |
| P(P14G)L6S* | 0.47 ± 0.02 | <0.0001b | >1000 |
| P(H15D)L6S* | 0.44 ± 0.06 | <0.0001b | >1000 |
acarried out in triplicate.
bbelow detection limit.
Oligomeric state of PL6S*
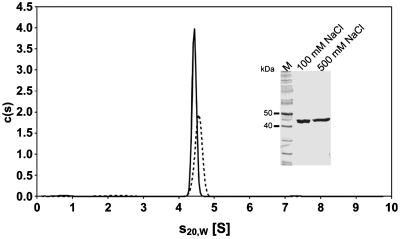
PvuII is a homodimer in solution (48). In order to investigate if the fusion with I-SceI has an effect on the oligomeric state, sedimentation velocity runs in the analytical ultracentrifuge were carried out. These experiments were done at high and low salt conditions. Under high salt conditions PL6S* sediments as a single species with a sedimentation coefficient of s20,w = 4.4 S (Figure 4). Increasing the protein concentration from 0.2 to 3.25 µM had no effect on the sedimentation coefficient (data not shown), indicating that PL6S* does not change its oligomerization state in this concentration range. Analysis of the diffusion broadening of the sedimenting boundary using the c(s) analysis in the program SEDFIT (45) yielded a molar mass of about 83 kg/mol. Since the molar mass of the monomer as calculated from amino acid composition is 46.7 kg/mol, the protein most probably is a homodimer. From the sedimentation analysis it can be ruled out, therefore, that the protein exists as a monomer in solution, since it sediments faster than a sphere of the corresponding mass. For the dimer a frictional ratio of f/f0=1.58 can be calculated from the sedimentation coefficient. As hydrated spherical proteins are expected to yield frictional ratios in the range of 1.1–1.2 (49), the shape of PL6S* must deviate substantially from a sphere, suggesting that it is an elongated homodimer and/or contains flexible regions that increase the frictional coefficient of the protein. Under low salt conditions the sedimentation coefficient increased slightly to s20,w = 4.6 S (Figure 4), and a molar mass of about 80 kg/mol was obtained from the c(s) analysis. This indicates that the PL6S* homodimer formed under low salt conditions is slightly more compact than under high salt conditions.
Figure 4.
Sedimentation coefficient distributions in an analytical ultracentrifugation experiment with PL6S* at different salt concentrations. PL6S* at concentrations of 3.3 or 3.5 µM was analyzed in sedimentation velocity experiments at 500 mM NaCl (solid line) or 100 mM NaCl (dashed line), respectively. c(s) analysis using the program SEDFIT yielded a single sedimenting species with s20,W = 4.4 S under high salt and s20,W = 4.6 S under low salt conditions, indicating that PL6S* is a stable homodimer. The purity and stability of the protein analyzed had been determined before by SDS-PAGE of 1.7 µg protein diluted in either low or high salt buffer (see insert). The theoretical molecular weight of one subunit of PL6S* is 46.7 kDa.
Identification of residues in PvuII that increase the specificity of the fusion enzyme
The fusion enzymes PL6S* and PL+S* are still able to cleave unaddressed PvuII-sites (Figure 3A), albeit with much lower rate than the addressed sites. There are in principle two possibilities to increase the preference of the fusion enzymes for the addressed site, which both rely on making the cleavage activity of the cleavage module PvuII dependent on the binding of the additional binding modules I-SceI* to the I-SceI site: (i) either by decreasing the activity (kcat, Km) of PvuII so that it cannot cleave DNA if not positioned by I-SceI*, or (ii) by weakening the dimer interface of PvuII to make a proper subunit–subunit interaction, which is a prerequisite for DNA cleavage, dependent on binding of the I-SceI* modules to their target sites.
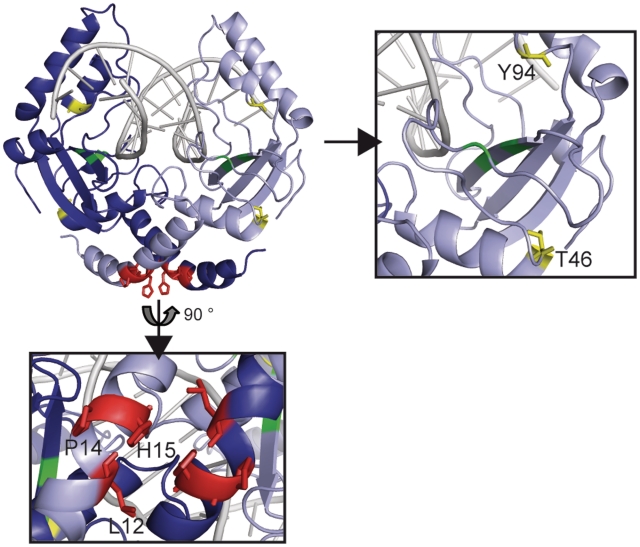
For the first approach (i) the mutations T46G, known to be responsible for high fidelity (Zhenyu Zhu, New England Biolabs, personal communication) and Y94F, known to have a decreased Mg2+ binding ability (50) were introduced into PvuII (Figure 5, right panel) and in the fusion enzyme. Indeed the PvuII(T46G,Y94F) variant showed an ∼100-fold decrease in activity compared to wtPvuII (data not shown). To weaken the PvuII dimer interface (ii) without affecting the catalytic activity too much we decided to choose residues in the N-terminal α-helices (Figure 5, left panel) which form close contacts between the two subunits and are far away from the active center (Figure 5, lower panel). The residues L12 and H15 were substituted by Glu and Asp, respectively, and thereby generate electrostatic repulsion between the two subunits. Furthermore the central residue of this helix P14 was substituted by Gly in order to affect the bending of the helix and by this reduce the interface contacts. These variants were introduced individually into wtPvuII and in the fusion enzyme as well. The PvuII(L12E), PvuII(P14G) and PvuII(H15D) variants were characterized with respect to their kinetic parameters, which showed, that the mutations had only a slight influence on the catalytic activity (Supplementary Table S1).
Figure 5.
Structural details of PvuII (pdb 1pvi). The catalytic centre composed of the residues D58, E68 and K70 is highlighted in green. The positions of substitutions introduced into PvuII are: T46G (yellow) increases the fidelity of PvuII; Y94F (yellow) has a reduced Mg2+ binding ability (50); L12E, P14G and H15D (red) are likely to weaken the dimer interface.
Characterization of fusion enzymes containing different amino acid substitutions in the cleavage module PvuII
We tested the effects of the amino acid substitutions T46G and Y94F in the fusion enzymes containing the L6 or the L+ linker by competition experiments with two internally labeled PCR fragments containing either the addressed site (S6P6S) or the unaddressed site (P) in equimolar concentration of enzyme and the two substrates (cPL6/+S* : cS6P6S: cP = 1:1:1) over 3 h. The fusion enzymes PL6S* (Figure 6A) and PL+S* (Figure 6C) containing wtPvuII as cleavage module cleave the addressed site by factors of ∼300 and 100 faster than the unaddressed site, respectively (Figure 6E and Table 4). Consistent with our finding that the PvuII(T46G, Y94F) variant has a highly reduced catalytic activity compared to wtPvuII, the variants P(T46G, Y94F)L6S* (Figure 6B) and P(T46G, Y94F)L+S* (Figure 6D) also have a much lower activity. However, they show only a 10–20-fold faster addressed than unaddressed cleavage rate (Figure 6E and Table 4).
Figure 6.
Competition cleavage experiments with fusion enzyme variants. The internally 32P labeled DNA fragments contain either the tripartite recognition sequence S6P6S (a) or a single PvuII site (u), at equimolar concentrations of 10 nM of both substrates. The two substrates were incubated with (A) PL6S*, (B) P(T46G, Y94F)L6S*, (C) PL+S* and (D) P(T46G,Y94F)L+S* for 3 h at 37°C; after defined time intervals samples were withdrawn from the reaction mixture and analyzed by gel electrophoresis. The cleavage product obtained by cleavage at the addressed tripartite recognition site is indicated by an arrow and the position of the expected cleavage product obtained by cleavage at the unaddressed PvuII site by an asterisk. (E) The autoradiograms (A–D) were quantified and the percentage of cleavage was plotted against time. Filled symbols show the addressed cleavage and open ones the unaddressed cleavage.
Since I-SceI is the part of the fusion enzymes which should be mainly responsible for specific DNA binding, we wanted to investigate, whether the affinity of the fusion enzymes to the tripartite recognition site (S6P6S) is higher than the affinity for a ‘stand-alone’ I-SceI recognition site (S) and is similar to the reported KD of 9.7 nM for I-SceI (51). For this purpose internally radiolabeled PCR fragments containing either the sites S6P6S or S were used for gel shift experiments under non-cleavage conditions. The affinity of the fusion enzymes for the S6P6S site was slightly lower than the affinity of I-SceI for its recognition site. A ‘stand-alone’ I-SceI site (S), however, is bound by the fusion enzyme with considerably lower affinity than the tripartite site (S6P6S) (Table 3). This indicates that PvuII is likely to contribute to the binding ability of the fusion enzyme to its specific site but disturbs the binding of I-SceI to a ‘stand-alone’ I-SceI site.
Table 3.
Summary of the binding constants KD determined for PL6S*, P(T46G,Y94F)L6S*, PL+S* and P(T46G,Y94F)L+S* on DNA substrates containing the tripartite site (S6P6S) or a single I-SceI site (S)
| Enzyme | Substrate | KD (nM) |
|---|---|---|
| PL6S* | S6P6S | 31 ± 3 |
| S | 144 ± 20 | |
| P(T46G,Y94F)L6S* | S6P6S | 31 ± 2 |
| S | 119 ± 16 | |
| PL+S* | S6P6S | 18 ± 1 |
| S | 212 ± 29 | |
| P(T46G,Y94F)L+S* | S6P6S | 31 ± 3 |
| S | >250 |
Optimizing the spacer length within the tripartite target sites with different distances between the PvuII and I-SceI sites
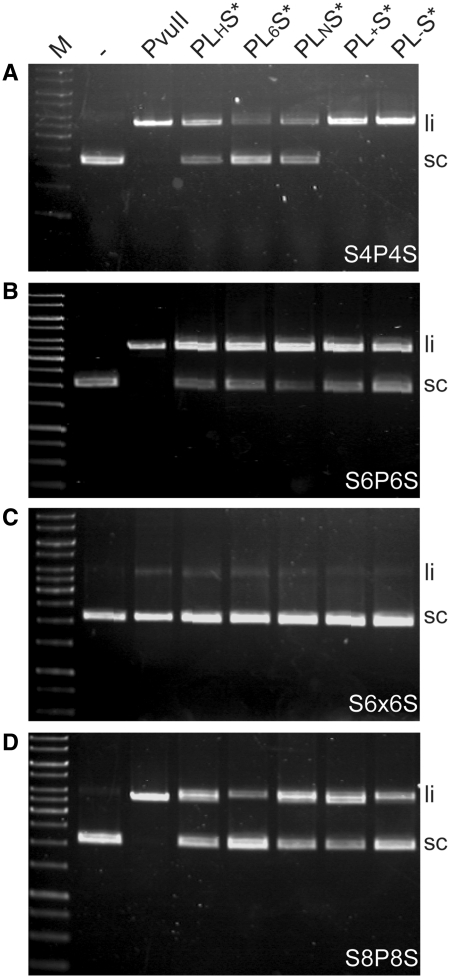
Before testing the PvuII variants with a proposed weakened dimer interface in the fusion constructs, we wanted to find out which linker is the preferred one in PvuII-I-SceI* fusion proteins for cleaving substrates with different distances between the PvuII site and the I-SceI site. For this, the linker variants PLHS*, PL6S*, PLNS*, PL+S* or PL-S* were assayed for cleavage activity on plasmid DNA substrates containing the target sites S4P4S, S6P6S, S6x6S or S8P8S (Table 2 and Figure 7). All variants cleave the addressed substrate with the 6 bp distance between I-SceI and PvuII site (Figure 7B) with nearly the same efficiency; in contrast, the substrate, which does not contain a PvuII target site (S6×6S) between the I-SceI sites, is not cleaved by any of the variants (Figure 7C). For the substrates with a decreased (S4P4S, Figure 7A) or increased (S8P8S, Figure 7D) distance between the I-SceI and PvuII sites differences in the cleavage efficiency were observed for the linker variants. The variants containing a charged linker (PL+S* or PL-S*) have a preference for the S4P4S substrate, PLHS* and PLNS* cleave the substrates with the following preference: S6P6S > S8P8S > S4P4S. PL6S* shows the preference predicted from the model (Figure 1B): S4P4S < S6P6S > S8P8S. For this reason the amino acid substitutions L12E, P14G and H15D were introduced only into PL6S*.
Figure 7.
DNA cleavage experiments with all linker variants and the different substrates at equimolar concentrations (8 nM). (A) S4P4S, (B) S6P6S, (C) S6x6S and (D) S8P8S were incubated with PvuII, PLHS*, PL6S*, PLNS*, PL+S* and PL-S*, respectively, for 3 h under optimized conditions (see main text).
Comparative analysis of the activity of PvuII-I-SceI fusion proteins in DNA cleavage assays
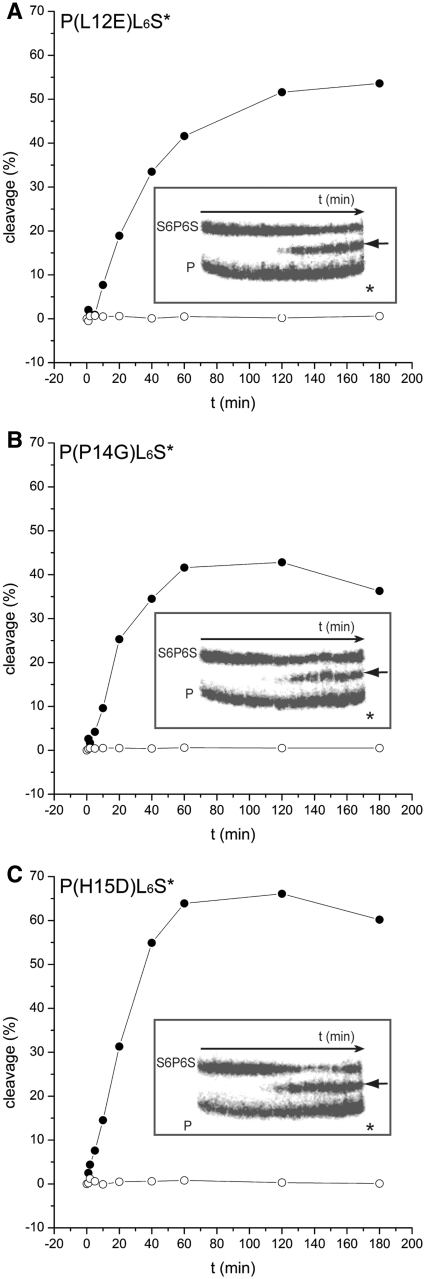
The specificity of these variants was tested and as expected from the kinetic parameters for PvuII(L12E), PvuII(P14G) and PvuII(H15D) (Supplementary Figure S1), P(L12E)L6S* (Figure 8A), P(P14G)L6S* (Figure 8B) and P(H15D)L6S* (Figure 8C) show a higher specific activity compared to P(T46G,Y94F)L6S* and P(T46G, Y94F)L+S* resulting in > 1000-fold faster addressed than unaddressed cleavage rate (Table 4). This preference is seen even at 10 times higher concentration of enzyme over the addressed and the unaddressed substrate (Supplementary Figure S2). It needs to be mentioned, however, that PL6S*, P(L12E)L6S*, P(P14G)L6S* and P(H15D)L6S* also attack substrates containing the dipartite site S6P, although much more slowly than the tripartite site S6P6S (Supplementary Figure S4). The preference for addressed sites could be even higher, since for none of them unaddressed cleavage could be detected, even after overnight incubation (data not shown), which makes these fusion proteins highly specific for the addressed substrate. We determined also the binding affinity of these variants for the S6P6S substrates. The KD-values obtained for P(L12E)L6S*, P(P14G)L6S* and P(H15D)L6S* were of the same magnitude as the KD of I-SceI (Table 5). On a substrate containing only an I-SceI site no specific binding could be detected (Supplementary Figure S3).
Figure 8.
Competition cleavage experiments with the fusion enzyme variants containing amino acid substitutions in the dimer interface and internally 32P labeled DNA fragments containing the addressed tripartite recognition site (S6P6S) or the unaddressed PvuII site alone (P). The experiment was performed as described in Figure 5. The autoradiogram is shown in the insert, the result of the quantification is shown as a reaction progress curve (percent cleavage vs. time): (A) P(L12E)L6S*, (B) P(P14G)L6S* and (C) P(H15D)L6S*. The arrow indicates the cleavage product obtained by addressed cleavage (closed circle) and the asterisk the position of the expected cleavage product of unaddressed cleavage (open circle).
Table 5.
Summary of the binding constants determined for P(L12E)L6S*, P(P14G)L6S* and P(H15D)L6S* on DNA substrates containing the tripartite site (S6P6S)
| Enzyme | Substrate | KD (nM) |
|---|---|---|
| P(L12E)L6S* | S6P6S | 9 ± 1 |
| P(P14G)L6S* | S6P6S | 9 ± 1 |
| P(H15D)L6S* | S6P6S | 2.3 ± 0.4 |
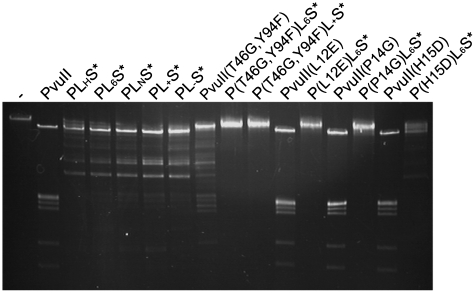
Since off-site target cleavage is a common problem of engineered nucleases of extended specificity (ZFN, meganucleases, etc.), we wanted to test to what extent our engineered fusion enzymes would cleave high molecular DNA at unwanted sites. For our fusion proteins PLxS* the most critical unwanted cleavage would be unaddressed cleavage i.e. cleavage at PvuII sites not addressed by I-SceI sites. For this purpose we used bacteriophage λ-DNA, which contains 15 PvuII sites, in a cleavage experiment (Figure 9). All variants produced, namely the different linker variants, the T46G, Y94F variants of PvuII alone or in the fusion enzyme context and the variants L12E, P14G and H15D also of PvuII alone or in the fusion enzyme context were used in a 1:1 ratio of enzyme over PvuII sites to see, if genomic DNA is a target for the meganuclease. The variants that we prepared at the beginning of our study (PLHS*, PL6S*, PLNS*, PL+S*, PL−S*) show cleavage of λ-DNA, even though not as complete as observed with wtPvuII. The variants with a decreased catalytic activity [P(T46G,Y94F)L6S*, P(T46G,Y94F)L+S]* show no cleavage of λ-DNA, but since PvuII(T46G,Y94F) also shows reduced cleavage activity compared to wtPvuII, the specific effect (i.e. absence of cleavage) is not as high as for P(L12E)L6S* and P(P14G)L6S* which also do not cleave λ-DNA, whereas PvuII(L12E) and PvuII(P14G) do so. Surprisingly, P(H15D)L6S* shows some cleavage, although it did not cleave the unaddressed substrate in the competition experiment (Figure 8C). We have verified the specificity of some of our fusion constructs also in vivo by demonstrating that E. coli cells producing fusion proteins are viable even in the absence of PvuII methyltransferase.
Figure 9.
DNA cleavage experiments with all fusion enzymes and all PvuII variants and bacteriophage λ-DNA, which harbors 15 PvuII sites. DNA and enzyme concentrations were chosen such, that the molar ratio between enzyme and PvuII sites is 1:1.
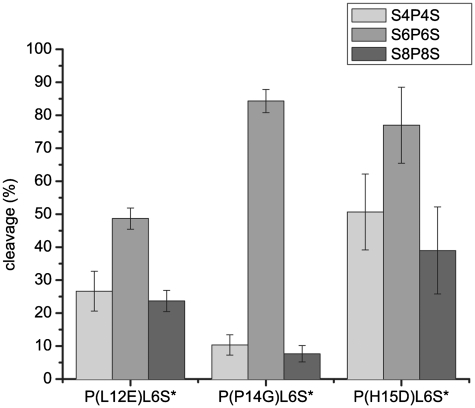
We were able to show that the variants P(L12E)L6S* and P(P14G)L6S* do not cleave PvuII sites on high molecular weight DNA and wanted to see finally, if these fusion enzymes have a preference for a certain distance between the I-SceI and PvuII sites. For this purpose, plasmid DNA containing either the S4P4S, S6P6S or S8P8S target was tested for cleavage by P(L12E)L6S*, P(P14G)L6S* and P(H15D)L6S*. The amount of linear DNA produced was determined by densitometry and the mean of three experiments was plotted (Figure 10). All enzymes cleave S6P6S preferentially, but to different degrees. Whereas P(L12E)L6S* has an ∼2-fold preference for S6P6S over S4P4s and S8P8S, and P(H15D)L6S* a 1.5- and 2-fold preference over S4P4S and S8P8S, respectively, P(P14G)L6S* shows a 8–10-fold preference for S6P6S over the other substrates. Taking all the results obtained together, P(P14G)L6S* is the fusion enzyme of choice, which cleaves highly preferentially the addressed substrate in the presence of unaddressed substrate, does not cleave unaddressed PvuII sites in λ-DNA and has a specificity for substrate with two I-SceI sites and a PvuII site in the middle separated by 6 bp on each site. These properties make this enzyme a suitable candidate to be tested for gene targeting in further in vivo experiments.
Figure 10.
Evaluation of DNA cleavage assays with three different plasmid substrates containing the sites S4P4S (light gray), S6P6S (gray) and S8P8S (dark gray). The variants P(L12E)L6S*, P(P14G)L6S* and P(H15D)L6S* were tested for cleavage of these substrates in equimolar concentrations of DNA and protein for 3 h at 37°C under optimized conditions. The amount of linear DNA was estimated by densitometry of the ethidium bromide stained agarose gel and the mean and standard deviation were calculated and plotted (n = 3).
DISCUSSION
Nucleases that are specific enough to recognize and cleave only unique DNA sequences in complex genomes are needed for the purpose of introducing new genetic material into complex genomes by homologous recombination. In gene targeting, new DNA sequences (e.g. complete genes) are introduced by making a specific double-strand cut at the locus of interest, which is repaired by recombination with the new DNA supplied in trans. There are two options to achieve this: (i) to use a nuclease that recognizes a sequence that had been specifically introduced into the genome at the locus of interest in a previous genome engineering experiment, or (ii) to reprogram a specific nuclease, or to program a non-specific nuclease to recognize a certain sequence at the locus of interest. We have pursued here the first option, namely to produce a nuclease with a unique specificity that could be used in particular for introducing new genes into genomes, in which recognition sequences for highly specific nucleases are already present or had been introduced for future targeted genome optimization as it has been done for example in agronomic crops (52,53).
Nucleases with extended specificity so far have been produced mainly by engineering homing endonucleases (‘meganucleases’) or by fusing a non-specific nuclease, typically the DNA cleavage domain of the FokI restriction endonuclease and three to four zinc fingers (→ZFN). In the new approach described here, we utilize the same architecture as utilized for ZFNs, triple helix forming oligonucleotide-linked nucleases, or TALENs, in which a DNA-binding module is fused to a DNA-cleavage module to generate an enzyme, whose distinct properties can be exploited to target a unique sequence in complex genomes. The binding module of choice for our purpose is derived from an inactive (denoted by *) variant of the monomeric homing endonuclease I-SceI (I-Sce*) that specifically recognizes an 18-bp sequence, but because of amino acid substitutions in the active site (Asp44→Ser or Asn; Asp145→Ala) is only competent in specific DNA binding but not in cleavage [cf. (20)]. In addition, I-SceI* lacks the nine C-terminal amino acid residues, which are not resolved in the co-crystal structure (40). For the DNA-cleavage module, we employ variants of the homodimeric type II restriction endonuclease PvuII. Different from the non-specific cleavage domain of FokI in the ZFNs, PvuII in our fusion proteins not only supplies the catalytic function, but also, in principle, could increase the overall specificity. Our PvuII-I-SceI* fusion proteins are homodimers (as shown by analytical ultracentrifugation), consisting of the central PvuII homodimer, whose subunits are fused at their C-termini to the N-termini of I-SceI*, which carries a C-terminal Strep-tag (Figure 1A). They bind the tripartite recognition site, consisting of a central PvuII recognition site and two flanking I-SceI recognition sites with higher affinity than a ‘stand-alone’ I-SceI recognition site.
A critical issue for the usefulness of engineered nucleases with extended specificity, in general, is that cleavage at so-called ‘off-target sites’ must be prevented (54). For PvuII-I-SceI* fusion proteins it is mandatory that they do not attack an unaddressed PvuII site (i.e. a PvuII site not flanked by I-SceI sites), and should prefer PvuII sites flanked on both sides with an I-SceI site over PvuII sites flanked on only one side with an I-SceI site, although we do not regard cleavage at PvuII sites flanked on only one side with an I-SceI site as critical, given the length of the combined recognition sequences of I-SceI and PvuII. We have employed a 2-fold strategy to achieve the required specificity of our fusion protein using either different linkers or introducing different amino acid substitutions into PvuII.
In a preliminary experiment, we had linked a catalytically inactive version of I-SceI via its N-terminus to the C-terminus of the single chain version of His6-tagged PvuII (scPvuII), in order to confirm that such a construct would not attack a DNA with one I-SceI site and without a PvuII site. To our surprise it turned out that the DNA was cleaved. Sequencing of the product identified the site of cleavage as a PvuII ‘star’ site (CAGCAG), which differs from a canonical PvuII site (CAGCTG) by a single base pair. Furthermore, at low Mg2+ (‘physiological’) concentrations the PvuII-I-SceI* fusion protein cleaves a plasmid DNA containing a composite site consisting of an I-SceI site and a PvuII site 8 bp downstream much better than scPvuII. These results showed that the scPvuII-I-SceI* fusion protein is functional and that I-SceI* can direct PvuII to a site that is related to a PvuII site. This result prompted us to pursue our experiments with fusion proteins consisting of wtPvuII and I-SceI*.
We had generated a structural model for PvuII-I-SceI* (Figure 1B). It suggested that the fusion protein designed to cleave a quasi-palindromic site consisting of a PvuII site flanked at a distance of ∼6 bp by two I-SceI sites oriented in opposite directions could be engineered with two linkers between the N-terminus of I-SceI* and the C-terminus of PvuII, ∼6–10 amino acid residues in length. This fusion protein would approach the DNA from one side. Of course, the distance of the PvuII and I-SceI sites and the linker length and its amino acid composition needed to be optimized. It turned out that a 6 bp distance and a linker of 6 amino acids (ASRTTG) proved to be optimal. A flexible 10 amino acid linker (ASTKQLVKSG) was also acceptable.
Since PvuII-I-SceI* fusion proteins even with optimized linkers were not absolutely specific for the addressed site, we introduced amino acid substitutions into PvuII that are known to affect Km and kcat. The intention was to increase the contribution of I-SceI* to specificity relative to PvuII, both in the ground (Km) and in the transition state (kcat). It is interesting to note that slowing down catalysis by introducing the T46G and Y94F amino acid substitutions into PL6S* and PL+S* results in a decrease of specificity [compare for example PL6S* ≡ 338 with P(T46G,Y94F)L6S*≡ 18]. This can be explained by an ‘allosteric coupling’ between the binding and the cleavage module in our fusion enzymes. A consequence of this coupling could be that the effect of reduced cleavage activity by the substitutions T46G and Y94F is only seen on the addressed site where PvuII is positioned correctly by the I-SceI mojety. On the unaddressed site this effect is not seen because the fusion enzyme is not able to bind in a catalytically productive manner. This is illustrated for example by our finding that PL6S* does not bind to an isolated PvuII site.
Since proper juxtaposition of the two subunits of PvuII is essential for double-strand cleavage we figured that interference with the subunit/subunit interface might affect PvuII activity but not that of the fusion protein, which is kept in a catalytically proficient dimer state by I-SceI* binding to its recognition sites. Although amino acid substitutions in the subunit/subunit interface did not abolish the DNA cleavage activity of PvuII, indicating that PvuII remains dimeric, they increased the specificity of the PvuII-I-SceI* fusion protein so much that no unaddressed DNA cleavage could be observed anymore. Presumably, the amino acid substitutions in the PvuII dimer interface impair DNA access for the catalytic center of PvuII in the fusion enzyme if the I-SceI/DNA interaction does not take place and thereby cannot support productive dimer formation of PvuII on the DNA.
In summary, given the detection limit of our DNA cleavage experiments, the preference of our best PvuII-I-SceI* fusion proteins is better than 1000 for an addressed site over an unaddressed site.
CONCLUSIONS
We have produced a fusion protein consisting of a LAGLIDADG homing endonuclease and a type IIP restriction enzyme for highly specific DNA cleavage as it is required for gene targeting. Nucleases with extended specificity that contain a DNA binding module fused to a DNA cleavage module invariably make use of the non-specific DNA cleavage domain of the type IIS restriction endonuclease FokI, as exemplified by the zinc finger nucleases. They have the advantage of being programmable by choosing appropriate zinc fingers to recognize series of trinucleotide sequences. The ZFN concept has been very successful, in particular after amino acid substitutions were introduced into the subunit-subunit interface in order to prevent homodimerization (55,56). Still, the problem of off-site cleavage, although very much reduced, still exists, as was systematically analyzed in two very recent studies (57,58).
We have wondered, whether other DNA cleavage modules could be used instead of the non-specific DNA cleavage domain of FokI to widen the repertoire of modules to be combined in engineering chimeric nucleases. In principle, also a specific DNA-cleavage module, for example a type IIP restriction endonuclease, could be used, as long as one can make DNA cleavage absolutely dependent on the binding of the DNA binding module. In this case one could take advantage of the specificity mediated by the cleavage module in addition to the DNA recognition by the DNA binding module. Our study shows that this is possible, as demonstrated for the tripartite site comprising the recognition sequences of the I-SceI homing endonuclease and the PvuII restriction endonuclease. To achieve the needed extraordinary specificity it is necessary to optimize the DNA binding module, the linker and the DNA cleavage module. Presumably, every fusion construct must be optimized individually. Our fusion enzyme is not freely programmable. However, for gene targeting purposes, one may introduce extended recognition sequences sites in the genome of interest at a so called ‘safe harbor’ locus, which can then be used reliably for genome engineering.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Material and Methods, Supplementary Table 1, Supplementary Figures 1–4 and Supplementary References [38 and 59].
FUNDING
Funding for open access charge: DFG (Deutsche Forschungsgemeinschaft) and the University of Giessen.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the expert technical assistance of Ms. Sabrina Stiehler.
REFERENCES
- 1.Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous Recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pingoud A, Wende W. Generation of novel nucleases with extended specificity by rational and combinatorial strategies. ChemBioChem. 2011;12:1495–1500. doi: 10.1002/cbic.201100055. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Kandavelou K, Chandrasegaran S. Custom-designed zinc finger nucleases: what is next? Cell. Mol. Life Sci. 2007;64:2933–2944. doi: 10.1007/s00018-007-7206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 7.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 8.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q. Rev. Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 9.Handel EM, Cathomen T. Zinc-finger nuclease based genome surgery: it's all about specificity. Curr. Gene Ther. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- 10.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 11.Galetto R, Duchateau P, Paques F. Targeted approaches for gene therapy and the emergence of engineered meganucleases. Expert Opin. Biol. Ther. 2009;9:1289–1303. doi: 10.1517/14712590903213669. [DOI] [PubMed] [Google Scholar]
- 12.Marcaida MJ, Munoz IG, Blanco FJ, Prieto J, Montoya G. Homing endonucleases: from basics to therapeutic applications. Cell. Mol. Life Sci. 2010;67:727–748. doi: 10.1007/s00018-009-0188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenschmidt K, Lanio T, Simoncsits A, Jeltsch A, Pingoud V, Wende W, Pingoud A. Developing a programmed restriction endonuclease for highly specific DNA cleavage. Nucleic Acids Res. 2005;33:7039–7047. doi: 10.1093/nar/gki1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon P, Cannata F, Concordet JP, Giovannangeli C. Targeting DNA with triplex-forming oligonucleotides to modify gene sequence. Biochimie. 2008;90:1109–1116. doi: 10.1016/j.biochi.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Tsai TL, Shieh DB, Yeh CS, Tzeng Y, Htet K, Chuang KS, Hwu JR, Su WC. The down regulation of target genes by photo activated DNA nanoscissors. Biomaterials. 2010;31:6545–6554. doi: 10.1016/j.biomaterials.2010.04.058. [DOI] [PubMed] [Google Scholar]
- 17.Kim YG, Chandrasegaran S. Chimeric restriction endonuclease. Proc. Natl Acad. Sci. USA. 1994;91:883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandavelou K, Mani M, Durai S, Chandrasegaran S. Engineering and applications of chimeric nucleases. In: Pingoud A, editor. Restriction Endonucleases. Berlin: Springer; 2004. pp. 413–434. [Google Scholar]
- 20.Lippow SM, Aha PM, Parker MH, Blake WJ, Baynes BM, Lipovsek D. Creation of a type IIS restriction endonuclease with a long recognition sequence. Nucleic Acids Res. 2009;37:3061–3073. doi: 10.1093/nar/gkp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fomenkov A, Too PH, Chan SH, Vaisvila R, Cantin BA, Mazzola L, Tam V, Xu SY. Targeting DNA 5mCpG sites with chimeric endonucleases. Anal. Biochem. 2008;381:135–141. doi: 10.1016/j.ab.2008.06.035. [DOI] [PubMed] [Google Scholar]
- 22.Chan SH, Bao Y, Ciszak E, Laget S, Xu SY. Catalytic domain of restriction endonuclease BmrI as a cleavage module for engineering endonucleases with novel substrate specificities. Nucleic Acids Res. 2007;35:6238–6248. doi: 10.1093/nar/gkm665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 24.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 25.Scholze H, Boch J. TAL effectors are remote controls for gene activation. Curr. Opin. Microbiol. 2011;14:47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl Acad. Sci. USA. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 30.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gingeras TR, Greenough L, Schildkraut I, Roberts RJ. Two new restriction endonucleases from Proteus vulgaris. Nucleic Acids Res. 1981;9:4525–4536. doi: 10.1093/nar/9.18.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 36.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 37.Colleaux L, d’Auriol L, Betermier M, Cottarel G, Jacquier A, Galibert F, Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986;44:521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- 38.Simoncsits A, Tjornhammar ML, Rasko T, Kiss A, Pongor S. Covalent joining of the subunits of a homodimeric type II restriction endonuclease: single-chain PvuII endonuclease. J. Mol. Biol. 2001;309:89–97. doi: 10.1006/jmbi.2001.4651. [DOI] [PubMed] [Google Scholar]
- 39.Tao T, and Blumenthal RM. Sequence and characterization of pvuIIR, the PvuII endonuclease gene, and of pvuIIC, its regulatory gene. J. Bacteriol. 1992;174:3395–3398. doi: 10.1128/jb.174.10.3395-3398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moure CM, Gimble FS, Quiocho FA. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J. Mol. Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 41.Gruen M, Chang K, Serbanescu I, Liu DR. An in vivo selection system for homing endonuclease activity. Nucleic Acids Res. 2002;30:e29. doi: 10.1093/nar/30.7.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joly E, Kirsch RD. An improved PCR-mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res. 1998;26:1848–1850. doi: 10.1093/nar/26.7.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClelland M, Hanish J, Nelson M, Patel Y. KGB: a single buffer for all restriction endonucleases. Nucleic Acids Res. 1988;16:364. doi: 10.1093/nar/16.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton J, editors. In Analytical Ultracentrifugation in Biochemistry and Polymer Science. England: Royal Society of Chemistry Cambridge; 1992. pp. 90–125. [Google Scholar]
- 47.Cheng X, Balendiran K, Schildkraut I, Anderson JE. Structure of PvuII endonuclease with cognate DNA. EMBO J. 1994;13:3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Athanasiadis A, Vlassi M, Kotsifaki D, Tucker PA, Wilson KS, Kokkinidis M. Crystal structure of PvuII endonuclease reveals extensive structural homologies to EcoRV. Nat. Struct. Biol. 1994;1:469–475. doi: 10.1038/nsb0794-469. [DOI] [PubMed] [Google Scholar]
- 49.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spyridaki A, Matzen C, Lanio T, Jeltsch A, Simoncsits A, Athanasiadis A, Scheuring-Vanamee E, Kokkinidis M, Pingoud A. Structural and biochemical characterization of a new Mg(2+) binding site near Tyr94 in the restriction endonuclease PvuII. J. Mol. Biol. 2003;331:395–406. doi: 10.1016/s0022-2836(03)00692-2. [DOI] [PubMed] [Google Scholar]
- 51.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 52.D’Halluin K, Vanderstraeten C, Stals E, Cornelissen M, Ruiter R. Homologous recombination: a basis for targeted genome optimization in crop species such as maize. Plant Biotechnol. J. 2008;6:93–102. doi: 10.1111/j.1467-7652.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 53.Yang M, Djukanovic V, Stagg J, Lenderts B, Bidney D, Falco SC, Lyznik LA. Targeted mutagenesis in the progeny of maize transgenic plants. Plant Mol. Biol. 2009;70:669–679. doi: 10.1007/s11103-009-9499-5. [DOI] [PubMed] [Google Scholar]
- 54.Petek LM, Russell DW, Miller DG. Frequent endonuclease cleavage at off-target locations in vivo. Mol. Ther. 2010;18:983–986. doi: 10.1038/mt.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 56.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 57.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 59.Huang Q, Quinones E. A spectroscopic method to determine the activity of the restriction endonuclease EcoRV that involves a single reaction. Anal. Biochem. 2008 doi: 10.1016/j.ab.2008.04.038. April 27 (doi:10.1016/j.ab.2008.04.038; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.