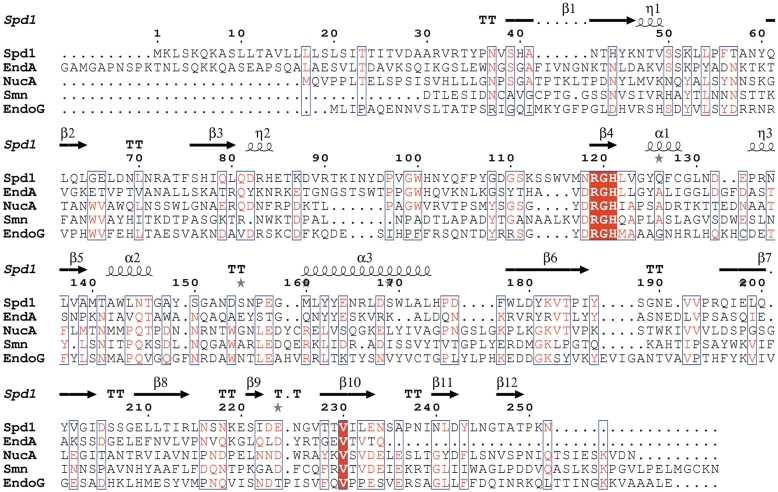
Figure 7.
Protein sequence alignment showing Spd1 in comparison to its closest known structural homologues: S. pyogenes nuclease (Spd1), pdb code 2xgr; S. pneumoniae nuclease (EndA), pdb code 3owv; Anabaena sp. nuclease (NucA), pdb code 1zm8; Drosophila melanogaster nuclease (EndoG), pdb code 3ism; Serratia marcescens nuclease (smn), pdb code 1smn. Secondary structure elements are taken from the Spd1 structure and indicated by the following, β-beta strand; α-alpha helix; TT-beta-turn; η-310 helix; grey star-residues modelled with an alternate conformation. Conserved residues are shown highlighted in red; areas of high homology are boxed in blue. Note Spd1 shows a variant asparagine residue at position 118, resulting in a NRGH catalytic motif, which was previously considered to be a conserved DRGH motif.