FIGURE 1:
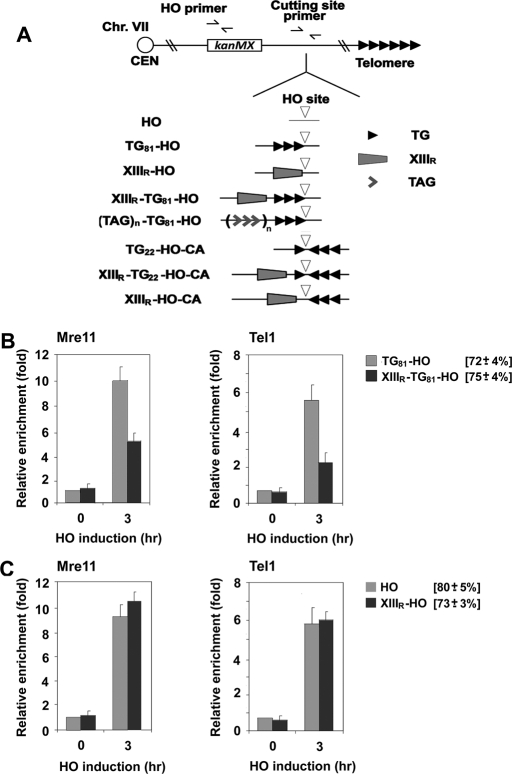
Effect of a subtelomeric sequence on localization of MRX and Tel1 to DNA ends with telomeric sequences. (A) Schematic of the HO cleavage site, the 81–base pair telomeric TG repeat, the XIII-R subtelomeric region, the 22–base pair telomeric TG repeat, and the TTAGGG repeats at the ADH4 locus on chromosome VII-L. In HO cells, the ADH4 locus was replaced with a DNA fragment containing the KanMX gene and an HO cleavage site (inverted triangle). The 81–base pair TG81 sequence (three repetitive arrowheads) or the XIII-R subtelomeric (XIIIR) sequence (trapezoid) was placed centromere-proximal to the HO cleavage site, generating the TG81-HO or XIIIR-HO strain, respectively. The XIIIR or TTAGGG repeat (wedge) sequence was integrated centromere-proximal to the TG repeat in TG81-HO cells, creating the XIIIR-TG81-HO or TAGn-TG81-HO strain, respectively. The 22–base pair TG22 (one arrowhead) sequence was inserted centromere-proximal to the HO cleavage site to generate the XIIIR-TG22-HO-CA or TG22-HO-CA strain. The inverted 81–base pair TG81 (CA) sequence was placed centromere-distal to the HO cleavage site for the XIIIR-TG22-HO-CA, TG22-HO-CA, or XIIIR-HO-CA strain. Centromere is shown as a circle on the left (CEN). The HO primer pair was designed to amplify a region 1 kbp away from the HO site, and the cutting-site primer pair was used to determine the HO cutting efficiency. (B) Effect of the chromosome XIIIR subtelomeric sequence on the association of Mre11 and Tel1 with DNA ends. TG81-HO (KSC2849) or XIIIR-TG81-HO (KSC2850) cells expressing myc-tagged Mre11 (Mre11-myc) and HA-tagged Tel1 (Tel1-HA) were transformed with the GAL-HO plasmid. Transformed cells were grown in sucrose and synchronized at G2/M with nocodazole. After arrest, galactose was added to the culture to induce HO expression. Aliquots of cells were collected at the indicated times after HO expression and subjected to ChIP assay to monitor the association of Mre11 or Tel1 with DNA ends. Coprecipitated DNA was analyzed by real-time PCR using the HO primer pair shown in A and the SMC2 primer pair for the control SMC2 locus on chromosome VI. Relative enrichment was determined from three independent experiments, and bars represent averages with standard deviations indicated by lines above. HO cutting efficiency for each construct is shown in square brackets. (C) Effect of the chromosome XIII-R subtelomeric sequence on the association of Mre11 and Tel1 with nontelomeric DNA ends. HO (KSC2851) or XIIIR-HO (KSC2852) cells expressing Mre11-myc and Tel1-HA were transformed with the GAL-HO plasmid, and association of Mre11 or Tel1 with DNA ends was analyzed by ChIP assay as in B.