Fig. 2.
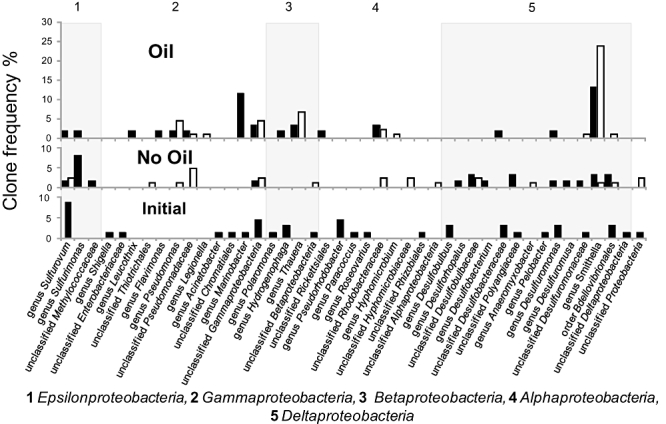
Phylogenetic affiliation of proteobacterial 16S rRNA sequences recovered from methanogenic microcosms. Clone frequency in bacterial 16S rRNA gene clone libraries from the inoculum and initial day 22 samples (bottom panel, total number of clones = 71) and in samples from methanogenic oil degrading microcosms (top panel, number of ‘day 302’ clones = 61, number of ‘day 686’ clones = 87) and control microcosms with no added oil (middle panel, number of ‘day 302’ clones = 62, number of ‘day 686’ clones = 87). Data from clone libraries from day 302 (filled bars) and day 686 (open bars) are shown. Clones were grouped into categories based on their genus, order, class or phylum level affiliation after phylogenetic analysis with the ARB software package using an RDP guide tree. The affiliation of individual sequences was cross-checked using the RDP taxonomical hierarchy with the Naïve Bayesian rRNA Classifier Version 2.0, July 2007.
