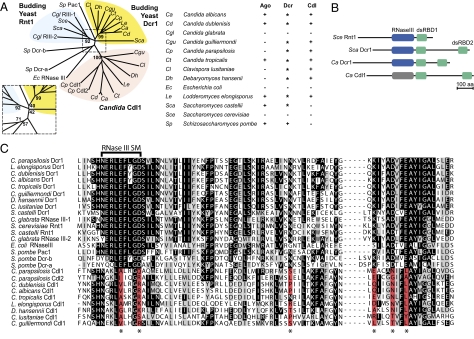
Fig. 1.
(A) Phylogenetic relationship between RNase III enzymes of indicated species. Bootstrap values for key nodes are shown, with central region (boxed) enlarged for clarity. Table indicates the presence (+) or absence (-) of the indicated gene. For Dicer, (*) designates budding-yeast Dicer, whereas + indicates canonical Dicer. (B) Domain structure of budding yeast RNase III enzymes. (C) Sequence alignment around the RNase III catalytic residues from the protein sequences from A. Sequence conservation is labeled in grayscale, with the most highly conserved residues most darkly colored. The six residues (corresponding to SceRnt1 E278, D282, N315, K346, D350, and E353) altered to catalytically inactive residues in the Cdl1 (Candida dicer-like) protein family are labeled (*) and shaded red when inactive. The 9-aa RNase III signature motif is labeled. ClustalW 1.81 with default setting was used to generate both the phylogenetic tree and the sequence alignment.