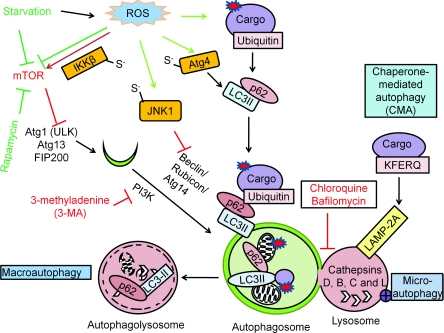
Figure 1. Autophagy mechanisms and regulation.
There are three different mechanisms of autophagy: (i) macroautophagy, the bulk degradation of cargo; (ii) microautophagy, involution of the lysosome around cargo; and (iii) chaperone-mediated autophagy (CMA), the degradation of the tagged cargo. The autophagosome and the lysosome fuse to degrade the cargo by lysosomal acidic hydrolases, including cathepsins D, B and L. Macroautophagy is regulated by starvation through the Atg1–Atg13 complex, which is inhibited by mTOR activation. Rapamycin or nutrient starvation causes mTOR inactivation, subsequent Atg1–Atg13 activation and activates macroautophagy. The beclin-1–PI3K complex is involved in autophagosomal expansion. 3-MA inhibits PI3K activity. Atg8–pro-LC3 is cleaved by Atg4, modified by PE to become LC3-II and inserted into the autophagosomes. The puncta formed by LC3-II and the electrophoretic separation of LC3-I and LC3-II are used as a marker for autophagosomal accumulation. RFP–GFP–LC3 (tfLC3) form puncta with both red and green fluorescence in the autophagosomes, and only red puncta when the autophagosomes fuse with lysosomes where GFP is inactivated, therefore tfLC3 is used to monitor autophagic flux. The degradation rate of long-lived proteins is another method for measurement of autophagic flux. p62 binds both LC3 and ubiquitin and therefore plays a role in targeting ubiquitinated proteins to the autophagosomes. Red arrows are indicative of negative regulators of autophagy, and green arrows are indicative of positive regulators of autophagy. Atg4 Cys81 thiol modification is involved in starvation and H2O2-induced autophagy. S-nitrosation of IKKβ and JNK1 are involved in NO-induced inhibition of autophagy.