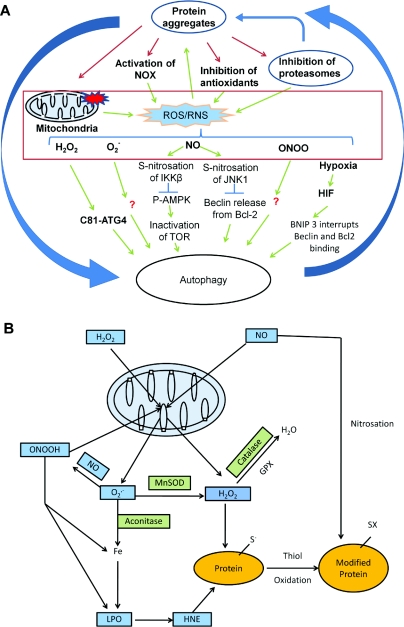
Figure 2. ROS/RNS production and signalling.
(A) ROS/RNS production and signalling of autophagy. ROS and RNS production can be induced by protein aggregates, generated by the mitochondrial respiratory activities and by other cellular oxidases. These ROS/RNS species can modify signalling molecules to stimulate or inhibit autophagy. Atg4 Cys81 thiol modification is involved in starvation and H2O2-induced autophagy. S-nitrosation of IKKβ and JNK1 are involved in NO-induced inhibition of autophagy. Although many ROS/RNS have effects on autophagy, besides a handful of known ROS/RNS-targeted autophagy regulators, the exact mechanisms for the effect of ROS/RNS on autophagy are largely unknown. (B) Mitochondrial production of ROS/RNS. Regardless of their sources, H2O2 and NO can target to the mitochondria. Mitochondrially produced superoxide (O2•−) can react with NO to produce peroxynitrite (ONOOH). O2•−, H2O2 and ONOOH can further damage the mitochondria in combination with iron and produce lipid peroxidation (LPO) products, including HNE. MnSOD, aconitase, GPX (glutathione peroxidase) and catalase are involved in generation and reduction of these products. HNE, NO and H2O2 can modify proteins to initiate signalling mechanisms or damage. The most sensitive moieties to oxidative modification are the thiols on cysteine residues. Redox reactions in protein modification are involved in signalling of autophagy.