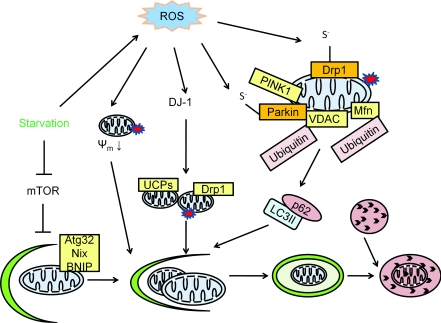
Figure 3. Mitophagy.
Mitophagy is the specific degradation of the mitochondria in response to global signals, including starvation and oxidative stress, or specific signals including mitochondrial targeting of signalling proteins or modification of mitochondrial proteins. In yeast, Atg32 is specific for mitophagy by targeting the mitochondria to the autophagosome. In mammalian cells, Nix is involved in mitochondrial clearance during erythrocyte maturation. In response to reduced cellular ATP, AMPK is activated and phosphorylates ULK1 and ULK2 (two Atg1 homologues) to activate both general macroautophagy and mitophagy. Parkinson's disease genes encoding α-synuclein, parkin, PINK1 and DJ-1 are all involved in mitophagy. A decrease in mitochondrial membrane potential (ψm) can be induced by ROS, and by targeting α-synuclein to the mitochondria. This decrease in mitochondrial membrane potential serves as a signal for mitophagy. In addition to a decrease in mitochondrial membrane potential, mitochondrial fission is another signal for mitophagy. PINK1 facilitates parkin targeting to the mitochondria, and ubiquitinates the mitochondrial outer membrane protein VDAC. Ubiquitinated VDAC can be recognized by p62 to initiate mitophagy. DJ-1 senses oxidative stress and serves a parallel pathway to maintain mitochondrial membrane potential and preserve mitochondria from fragmentation. Many of the regulators of mitophagy can be regulated by ROS. For example, α-synuclein is nitrated and, as a consequence, increases its aggregation propensity. Parkin can be sulfonated and S-nitrosated. Drp1 S-nitrosation is also involved in regulation of mitochondrial fission and associated induction of mitophagy.