FIGURE 1.
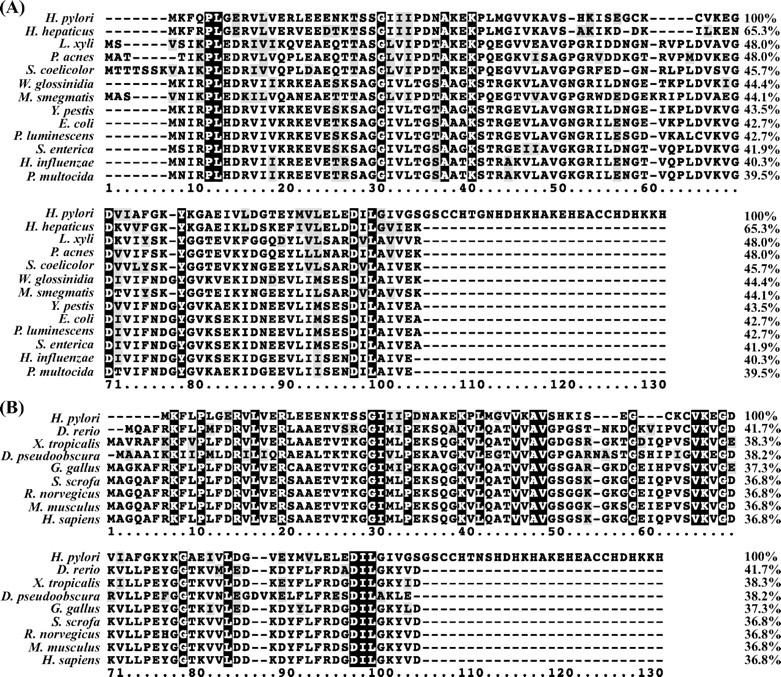
Sequence alignment of H. pylori HspA with homologues from other pathogenic microorganisms (A) or model species (B). Identical residues are marked with a black background, and conserved ones are shaded in light gray. The sequence alignment was done by ClustalW (version 1.83), and the sequence similarities were calculated by the Needle program from the EMBOSS package (version 3.0) using the H. pylori homologue as the template. The following sequences were retrieved from the GenBank™ Protein Databank. A, HspA (H. pylori; AAP51175.1), co-chaperonin GroES (Helicobacter hepaticus; NP_860731), co-chaperonin GroES (Leifsonia xyli xyli; YP_062808), 10-kDa chaperonin (Propionibacterium acnes; YP_056460), co-chaperonin GroES (Streptomyces coelicolor; NP_628919), GroES (Wigglesworthia glossinidia; AAK07426), chaperonin GroS (Mycobacterium smegmatis; YP_885961), co-chaperonin GroES (Yersinia pestis; NP_403998), co-chaperonin GroES (E. coli; NP_418566.1), co-chaperonin GroES (Photorhabdus luminescen; NP_931325), co-chaperonin GroES (Salmonella enterica; YP_219195), co-chaperonin GroES (Haemophilus influenzae; NP_438700), and co-chaperonin GroES (Pasteurella multocida; NP_246043). B, heat shock 10-kDa protein (Sus scrofa; AAP32465), heat shock 10-kDa protein (Homo sapiens; NP_002148), chaperonin 10 (Rattus norvegicus; AAC53361.1), chaperonin 10 (Mus musculus; AAF67345.1), chaperonin 10 (Gallus gallus; NP_990398.1), chaperonin 10 (Xenopus tropicalis; AAH77653.1), chaperonin 10 (Danio rerio; AAH71419.1), GA10877-PA (Drosophila pseudoobscura; EAL31011.1), and HspA (H. pylori; AAP51175.1).
