Abstract
Embryonic stem (ES) cells are pluripotent-undifferentiated cells that have a great interest for the investigation of developmental biology. Murine ES cells maintain their pluripotency by the supplementation of the leukemia inhibitory factor (LIF). LIF is reported to act as a matrix-anchored form, and immobilized cytokines are useful to sustain their signaling on target cells. In this study, we used the immobilizable fusion protein composed of LIF and IgG-Fc region, which was used as a model of the matrix-anchored form of LIF to establish a novel system for ES cell culture and to investigate the effect of immobilized LIF on maintenance of ES cell pluripotency. Mouse ES cells maintained their undifferentiated state on the surface coated with LIF-Fc. Furthermore, when cultured on the co-immobilized surface with LIF-Fc and E-cadherin-Fc, mouse ES cells showed characteristic scattering morphologies without colony formation, and they could maintain their undifferentiated state and pluripotency without additional LIF supplementation. The activation of LIF signaling was sustained on the co-immobilized surface. These results indicate that immobilized LIF and E-cadherin can maintain mouse ES cells efficiently and that the immobilizable LIF-Fc fusion protein is useful for the investigation of signaling pathways of an immobilized form of LIF in the maintenance of ES cell pluripotency.
ES3 cells are pluripotent-undifferentiated cells derived from the inner cell mass of a blastocyst, and they retain the potentiality of multilineage differentiation in vitro and self-renewal activity (1, 2). For the maintenance of pluripotency, mouse ES cells are cultured on a feeder layer of mouse embryonic fibroblasts (MEF) or incubated with LIF that is produced by MEF. LIF is a pleiotropic cytokine, which induces the differentiation of leukemia cell lines into macrophages (3) and the expression of acute phase proteins in hepatocytes (4), as well as inhibits proliferation of endothelial cells (5).
Several studies suggest that the biological signals of growth factors and cytokines are mediated by two different forms, the secreted form and the cell membrane- or matrix-anchored forms, which stimulate different signal transduction cascades (6–8). It is reported that the LIF signal is mediated by either a soluble form or by a form bound to extracellular matrices, both of which induce different effects on cells (9, 10).
Recently, we reported the application of immobilizable fusion proteins for the analysis of cell function, and we also reported the establishment of a culture system for stem cells (11–13). When cells were cultured on the fusion protein of E-cadherin and IgG Fc domain (E-cad-Fc), they showed scattering behavior and high proliferative ability (11). We also reported the novel method of immobilizing epidermal growth factor as a juxtacrine model, and we applied this system to the analysis of the function of immobilized epidermal growth factor on hepatocytes (13).
In this study, we focused on the effect of the immobilized form of LIF on the maintenance of ES cell features, and we used LIF and IgG-Fc region to design the immobilizable fusion protein (LIF-Fc). By using this model protein, we investigated the effect of immobilized LIF on maintenance of ES cell pluripotency. Furthermore, to establish an effective culture system of mouse ES cells, we applied the co-immobilized surface of E-cad-Fc and LIF-Fc.
EXPERIMENTAL PROCEDURES
Cell Culture—We used the following mouse ES cell lines: two feeder-dependent ES cell lines, Jxl1 cells (established from 129/SvJ x 129/SvImJ mice) and R1 cells (14); and feeder-free EB3 cells (15). Jxl1 and R1 cells were maintained on mouse embryonic fibroblasts in Dulbecco's modified Eagle's medium (Chemicon), supplemented with 1% nonessential amino acids (Chemicon), 0.1 mm β-mercaptoethanol (Sigma), 15% (v/v) fetal bovine serum (FBS), and 1,000 units/ml LIF. For several experiments, Jxl1 and R1 cells were adapted to the feeder-free culture condition. Cells were seeded in the media containing 15% knock-out serum replacement (Invitrogen) and 5,000 units/ml LIF. EB3 cells were maintained on 0.1% gelatin-coated surfaces in knock-out/Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 1 mm l-glutamine, 1% nonessential amino acids (Invitrogen), 0.1 mm β-mercaptoethanol (Sigma), 20% (v/v) FBS, and 2,000 units/ml ESGRO (Chemicon). Cells were passaged every 2 or 3 days with 0.25% trypsin, 1.0 mm EDTA solution (Invitrogen). M1 (D+) cells (provided by Cell Resource Center for Biomedical Research, Tohoku University) were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% (v/v) FBS. All media contained 50 μg/ml penicillin, 50 μg/ml streptomycin, and 100 μg/ml neomycin.
Construction and Purification of Fusion Proteins—Expression and purification of E-cad-Fc fusion proteins were described previously (16). In brief, the E-cadherin extracellular domain cDNA, which was generated from mouse E-cadherin full-length cDNA provided by the RIKEN BRC DNA Bank (code 1184), and mutated mouse IgG1-Fc domain cDNA (T252M/T254S), which have high affinity to protein A, were ligated with pRC/CMV (Invitrogen) fragment to generate the expression vector “pRC-ECFC.” Using Lipofectamine reagent (Invitrogen) according to the manufacturer's directions, CHO-K1 cells were transfected with pRC-ECFC. After selection of a highly expressing clone (4G7) with 400 μg/ml G418 (Invitrogen), conditioned media were collected.
To construct LIF-Fc, the cDNA that encodes mouse LIF was amplified with KOD plus DNA polymerase (TOYOBO) from the cDNA of mouse embryonic fibroblasts. The specific primer pair for mouse LIF was used for amplification: 5′-AAG CTT CAT AAT GAA GGT CTT GGC CG-3′ and 5′-GCG GCC GCT GAA GGC CTG GAC CAC CAC AC-3′; the underlining represents the HindIII and NotI recognition sites, respectively. To generate pRC-LIF-Fc vector, pGEM-T easy vector (Pro-mega) containing mouse LIF cDNA was digested by HindIII and NotI and subcloned into pRC/CMV vector containing mutated mouse Fc fragment cDNA (16). CHO-K1 cells were transfected with pRC-LIF-Fc vector, and a highly expressing clone was selected as mentioned above.
The fusion proteins were loaded onto a rProtein A FF column (GE Healthcare). The column was washed with 20 mm phosphate buffer (pH 7.0), and the bound proteins were eluted using 0.1 m sodium citrate (pH 2.7) followed by neutralization with a 1/5 volume of 1.0 m Tris-HCl (pH 9.0). Eluates were dialyzed for 3 days in PBS containing 0.9 mm CaCl2 and 0.9 mm MgCl2.
To prepare the coated surface with E-cad-Fc and/or LIF-Fc, purified E-cad-Fc and LIF-Fc solution was directly added to nontreated polystyrene plates at the indicated concentration. After 2 h of incubation at 37 °C, plates were washed with PBS once, and then cells were seeded.
Western Blot Analysis—The total cellular protein was extracted with lysis buffer (20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Nonidet P40, 1% Triton X-100, 1 mm Na3VO4, 1 mm NaF, and proteinase inhibitors), and cell lysates were centrifuged at 17,860 × g for 15 min. Protein concentration of the supernatant was measured using a DC protein assay kit (Bio-Rad), and samples were separated by electrophoresis on 7.5% polyacrylamide gels and electrophoretically transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore). Blots were probed with anti-murine LIF antibody (Chemicon), anti-STAT3 antibody (BD Biosciences), anti-phospho-STAT3 (Tyr-705) antibody (Cell Signaling Technology), anti-p44/42 MAPK antibody (Cell Signaling Technology), anti-LIF receptor antibody (Santa Cruz Biotechnology), anti-gp130 antibody (Santa Cruz Biotechnology), or anti-phospho-p44/42 MAPK (Thr-202/Tyr-204) antibody (Cell Signaling Technology), followed by horseradish peroxidase-conjugated secondary antibodies, and developed by ECL reagent (GE Healthcare) or Immobilon Western Chemiluminescent horseradish peroxidase substrate (Millipore).
Definition of LIF-Fc Activity—To determine the specific activity of purified LIF-Fc, we analyzed the ability to induce the differentiation of M1 cells (17). M1 cells were suspended in Dulbecco's modified Eagle's medium with a final concentration of 20% FBS, 0.3% agar (Dojindo Molecular Technologies) and LIF samples, and then they were seeded into 24-well plates at the concentration of 75 cells/well. ESGRO (Chemicon) was used as a control to determine LIF activity. Cultures were incubated for 10 days in a fully humidified atmosphere at 5% CO2. Cultures were scored using a microscope at ×40 magnification, scoring as differentiated any colonies with a corona of dispersed cells or composed wholly of dispersed cells. A standard of 50 units was defined as the concentration that induces the differentiation of 50% of M1 colonies.
Characterization of Immobilized Proteins—Following the manufacturer's instructions, purified LIF-Fc was labeled with HiLyte Fluor™ 555 labeling kit-NH2 (excitation, 555 nm; emission, 570 nm; Dojindo). Labeled LIF-Fc (555-LIF-Fc) solution was diluted 1:10 with native LIF-Fc solution when the concentration of total LIF-Fc was greater than 1.0 μg/ml, so that the fluorescence intensity is a linear function of labeled protein concentration. 555-LIF-Fc, native LIF-Fc, and E-cad-Fc were diluted with PBS to the designated concentration, and then 50 μl of mixtures were added to 96-well polystyrene plates. After 2 h of incubation at 37 °C, the fluorescence intensity of nonadsorbed fraction was measured with microplate reader (excitation, 535 nm; emission, 595 nm). The amount of immobilized LIF-Fc was estimated from a calibration curve prepared with 555-LIF-Fc and native LIF-Fc.
Adhesion and Growth Assays—Cells were seeded at a density of 3.0 × 104 cells/well into 96-well plates precoated with the indicated substrate. After 4 h of culture, medium and nonadherent cells were removed, and cells were washed with the culture medium. Adherent cells were stained with Alamar Blue reagent (BIOSOURCE), and absorbance at 570 nm was measured using a microplate reader. For the cell growth assay, cells were seeded at a density of 500 cells/well into a 96-well plate coated with the indicated substrate. The cell number was evaluated at 5 days after seeding.
ELISA—The amount of remaining LIF-Fc after the cultivation of ES cells was analyzed by ELISA method. ES cells were seeded onto the surface coated with E-cad-Fc and LIF-Fc and cultured for 3 days. To remove the adhered ES cells, we incubated them in a 2 mm EDTA/PBS solution for 30 min at 37 °C. After washing three times with PBS, plates were incubated for 16 h with 1.0% bovine serum albumin (BSA)/PBS solution to block unspecific interaction. The remaining LIF-Fc was detected after incubation with an anti-murine LIF antibody for 2 h, followed by peroxidase-conjugated secondary antibody for 1 h. We used 3,3′,5,5′-tetramethylbenzidine (TMB) as a substrate for peroxidase, and we measured the absorbance at 450 nm.
The amount of adsorbed LIF-Fc onto the gelatinized or E-cad-Fc-coated surface was also analyzed by ELISA. The media containing 1,000 units/ml LIF or LIF-Fc were added to the gelatinized or E-cad-Fc-coated surface. After 3 days of incubation at 37 °C, the amount of nonadsorbed LIF or LIF-Fc in the supernatant was analyzed by Quatikine Mouse LIF immunoassay kit (R & D Systems). The amount of adsorbed LIF or LIF-Fc was measured by the same method as mentioned above.
ALP Staining and Immunofluorescence—Alkaline phosphatase activity was determined using a Sigma Diagnostics alkaline phosphatase (ALP) kit (Sigma). For immunofluorescence staining, cells were fixed with 8% formaldehyde solution (pH 7.0–7.5; Wako Pure Chemical) for 10 min and permeabilized with 0.2% Triton X-100 for 2 min at room temperature. Fixed cells were incubated with Image-iT FX signal enhancer (Invitrogen) for 30 min at room temperature. Oct-3/4 was stained with an anti-mouse Oct-3/4 polyclonal antibody (H-134; Santa Cruz Biotechnology) for 2 h followed by an Alexa Fluor 546-conjugated secondary antibody (Invitrogen) for 1 h. Nuclei were counterstained with 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Samples were observed by fluorescence microscopy. The ratio of Oct-3/4-positive cells was calculated by ImageJ software.
RT-PCR Analysis—Total RNA was isolated with TRIzol reagent (Invitrogen). The first strand cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Invitrogen), and PCR was carried out with rTaq polymerase (TOYOBO) in the reaction buffer containing 1.5 mm MgCl2. Primers used are listed in Table 1. PCR products were analyzed by 2% agarose gel electrophoresis.
TABLE 1.
PCR primers used in this study
| Gene (Unigene symbol) | 5′ primer (5′ to 3′) | 3′ primer (5′ to 3′) | Product length | Annealing temperature | Cycle no. |
|---|---|---|---|---|---|
| bp | °C | ||||
| Oct-3/4 (Mm.17031) | GAAGTTGGAAAGGTGGAACC | GCCTCATACCTTCTCCGTTGG | 528 | 60 | 20 |
| TCTTTCCACAGGCCCCCGGCTC | TGCGGGCGGCATGGGGAGATCC | 224 | 60 | 20 | |
| Zfp42/Rex-1 (Mm.285848) | AAAGTGAGATAGCCCCGAG | TCCCATCCCTTCAATAGCA | 930 | 60 | 20 |
| ACGAGTGGCGTTTCTTCTTGGGA | TATGACTCATTCCAGGGGGCACT | 287 | 60 | 20 | |
| Nanog (Mm.6047) | GAGGAAGCACGAATTCTGG | AAGTTATGGGCGGAGCAGC | 710 | 60 | 20 |
| GCGGCTCACTCCTTCTGACTT | GACCAGGAAACCCACACTCAT | 163 | 60 | 20 | |
| Wnt3 (Mm.159091) | TAGAGCTAGCTCCGGGCGATGA | TTGCCTTAACAAGACCACGAAA | 297 | 62 | 20 |
| Actcl (Mm.686) | CCAGATCATTTTGAGACCTTCAA | GAACATTATAGTTACACCATCGC | 124 | 59 | 30 |
| Nkx2.5 (Mm.41974) | GAGCCTACGTGACCCTGACCCAG | TGACCTGCGGGACGTGAGCTTCA | 264 | 60 | 30 |
| Gata4 (Mm.247669) | CTGGAGGCGGATGGGACGGGACACTAC | CCGCAGGCATACATACAGGCTCACC | 207 | 62 | 24 |
| α-Fetoprotein (Mm.358570) | TCGTATTCCACAGGAGG | AGGCTTTTGTTCACCAG | 173 | 55 | 30 |
| Bmp2 (Mm.103205) | GGGACCCGCGTCTTCTAGTGTTGC | TGAGTGCCTCGGTACAGATCTAGCA | 249 | 60 | 30 |
| Polr2a (Mm.16533) | CTGATGCGGTGCTGAGTGAGAAGG | GCGGTTGACCCATGACGAGTG | 237 | 60 | 24 |
| Gapdh (Mm.333399) | CTCATGACCCAGTCCATGC | CTCTTGCTCGTGTCCTTGC | 532 | 60 | 20 |
Tetraploid Aggregation—Embryos were generated from ES cells by aggregating them with four-cell stage tetraploid CD-1 embryos generated by electrofusion as described previously (18, 19). ES cell:embryo aggregates were cultured overnight, and those that formed blastocysts were transferred to the uteri of pseudo-pregnant females surrogate mothers. Embryos were allowed to develop in utero before harvesting for analysis. The Animal Care Committee of the Medical College of Wisconsin approved all animal procedures used in this study.
RESULTS
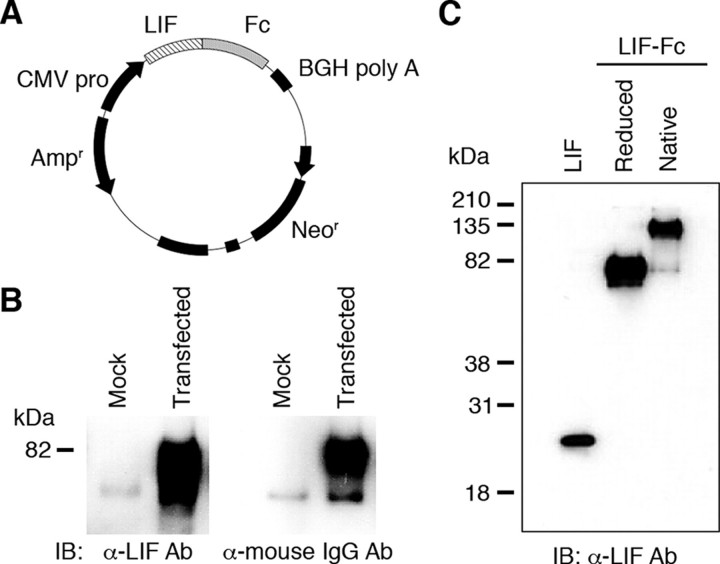
Construction, Purification, and Characterization of LIF-Fc—CHO-K1 cells were transfected with an expression vector for LIF-Fc (Fig. 1A), and G418-resistant clones were selected. A highly expressing clone was selected by ELISA (data not shown). LIF-Fc was purified from the conditioned media of the selected clone and analyzed by Western blotting. Under reduced conditions, the LIF-Fc was detected as an ∼75-kDa single band that was recognized by both anti-LIF antibody and anti-mouse IgG antibody (Fig. 1B). Under nonreduced conditions, LIF-Fc gave rise to an ∼150-kDa single band, indicating that LIF-Fc forms the homodimers (Fig. 1C).
FIGURE 1.
Construction and expression of the fusion protein of LIF-Fc. A, plasmid vector to produce LIF-Fc was constructed as described under “Experimental Procedures.” B, expression of fusion protein was checked by Western blotting. IB, immunoblot; Ab, antibody. The fusion protein was detected by either an anti-mouse IgG antibody or anti-LIF antibody. C, LIF-Fc was separated by SDS-PAGE under a reduced or native condition.
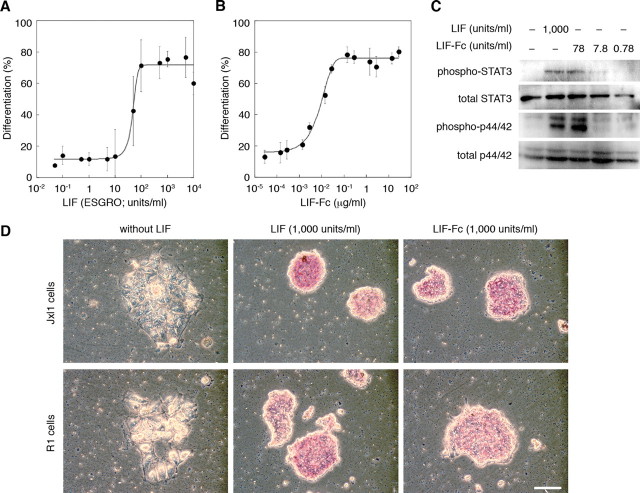
Analysis of LIF-Fc Activity—The activity of purified LIF-Fc was defined as an ability to induce differentiation of M1 cells (Fig. 2, A and B). LIF-Fc induced the differentiation of M1 cells in a dose-dependent manner, and the specific activity of LIF-Fc was estimated as described under “Experimental Procedures” (in this case 5,074 units/μg).
FIGURE 2.
The activity of LIF (A) and soluble LIF-Fc (B) was measured by M1 assay, as described under “Experimental Procedures.” The ability of soluble LIF-Fc to maintain an undifferentiated state of mouse ES cells was determined by the phosphorylation of STAT3 and MAPK (C) and by ALP activity (D). Bar indicates 100 μm.
The activity of LIF-Fc was also evaluated by the ability to activate the STAT3 and MAPK signaling pathways in mouse ES cells. As shown in Fig. 2C, LIF-Fc also activated these pathways, similar to the activation induced by recombinant LIF (ESGRO). ALP activity is a marker of the undifferentiated state of ES cells. The ability of LIF-Fc to maintain the undifferentiated state of mouse ES cells was analyzed by ALP activity. Mouse ES cells maintained ALP activity by LIF-Fc supplementation (Fig. 2D), indicating that LIF-Fc could retain the original activity of LIF, even though LIF is conjugated with the Fc domain of IgG and forms homodimers.
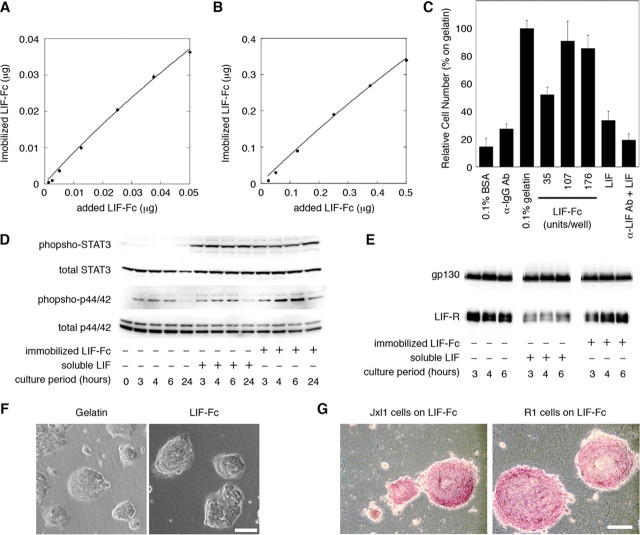
Adsorption of LIF-Fc on Polystyrene Surface and Effect of an Immobilized LIF-Fc on ES Cells—First, the amount of LIF-Fc adsorption onto a polystyrene surface was estimated by measuring the fluorescence intensity of 555-LIF-Fc. Adsorption of LIF-Fc to the polystyrene surface increased in a dose-dependent manner, and the curve could be approximated to Langmuir's adsorption isotherm (Fig. 3, A and B). The amount of immobilized LIF-Fc onto polystyrene surface was estimated as about 67 ± 15% of initial amount.
FIGURE 3.
The amount of immobilized LIF-Fc was measured at a low concentration (A) and a high concentration (B) of LIF-Fc. The data indicate means ± S.E. of five separate experiments. C, adhesion of mouse ES cells onto the surface of 96-well plate coated with 0.1% BSA, anti-mouse IgG antibody, 0.1% gelatin, LIF-Fc, recombinant LIF (10,000 units/ml), or anti-LIF antibody followed by recombinant LIF (10,000 units/ml). After 3 h of incubation, ES cells adhered to a LIF-Fc-coated surface with equivalent efficiency as to a 0.1% gelatin-coated surface. The data indicate means ± S.D. of three separate experiments. D, activation of STAT3 and MAPK by immobilized LIF-Fc was analyzed by Western blotting. The phosphorylation of MAPK was sustained as long as 24 h after culturing. E, internalization of the LIF receptor and gp130 was analyzed by Western blotting. The amount of LIF receptor did not change on the LIF-Fc immobilized surface. F, morphological observation of R1 cells cultured on various substrates. G, ALP activity of ES cells cultured for 4 days in the 24-well plate immobilized with LIF-Fc. ES cells formed aggregated colonies on LIF-Fc-coated surface, and they maintained ALP activity without LIF supplementation. Scale bar indicates 100 μm.
Next, we tested the effect of immobilized LIF-Fc on ES cell culture. EB3 cells adhered to the LIF-Fc-coated surface but not to the surface coated with mouse IgG (Fig. 3C), indicating that cell adhesion is mediated by immobilized LIF-Fc not by the Fc region. In addition, to clarify the effect of the Fc region on the stable adsorption of LIF-Fc protein, we directly coated native LIF to the polystyrene surface. ES cells did not adhere to the surface coated with native LIF; therefore, the Fc region is essential for the stable adsorption of LIF, and LIF-Fc could act as a substrate for adhesion of ES cells. On the LIF-Fc-coated surface, ES cells formed tightly aggregated colonies (Fig. 3F); similar morphology is observed on the conventional gelatin-coated surface. To ascertain whether ES cells maintain their undifferentiated phenotypes on immobilized LIF-Fc, we examined the activity of STAT3 and MAPK. STAT3 was activated on the LIF-Fc immobilized surface at the same level as on the gelatinized surface with LIF, and the activation of STAT3 was sustained to 24 h. The activation of MAPK on the LIF-Fc-coated surface was slightly higher than on a conventional gelatinized surface with LIF supplementation (Fig. 3D). When activated by soluble ligands, many cytokine receptors are usually incorporated into cells to control the strength and period of signals. The LIF receptor is also internalized and down-regulated (20). Within 3 h after stimulation, the protein level of the LIF receptor was decreased by the stimulation of soluble LIF, although there were no differences on the level of gp130 as described previously (Fig. 3E) (20). On the other hand, immobilized LIF-Fc had no effect on the incorporation of the LIF receptor, indicating that immobilized LIF would not be incorporated into the cells. Furthermore, ES cells grown on the LIF-Fc-coated dishes formed aggregated colonies (Fig. 3F) and showed high ALP activity (Fig. 3G). These results indicate that ES cells could maintain their undifferentiated phenotypes on a LIF-Fc-coated surface.
Maintenance of ES Cell Features on the Co-immobilized Surface of LIF-Fc and E-cad-Fc—We demonstrated that ES cells could maintain their undifferentiated phenotypes on a LIF-Fc-coated surface without additional LIF supplementation; however, they still form tight aggregated colonies. Furthermore, their adhesion to LIF-Fc was not strong; therefore, they could easily detach from the surface and form floating cell aggregates, like embryoid bodies (data not shown). Several reports suggested that aggregation would induce heterogeneous signals depending on the position of cells (21–23). To clear these problems, we used the co-immobilized surface coated with LIF-Fc and the fusion protein of E-cadherin and Fc fragment (E-cad-Fc) that could maintain ES cells without colony formation (11).
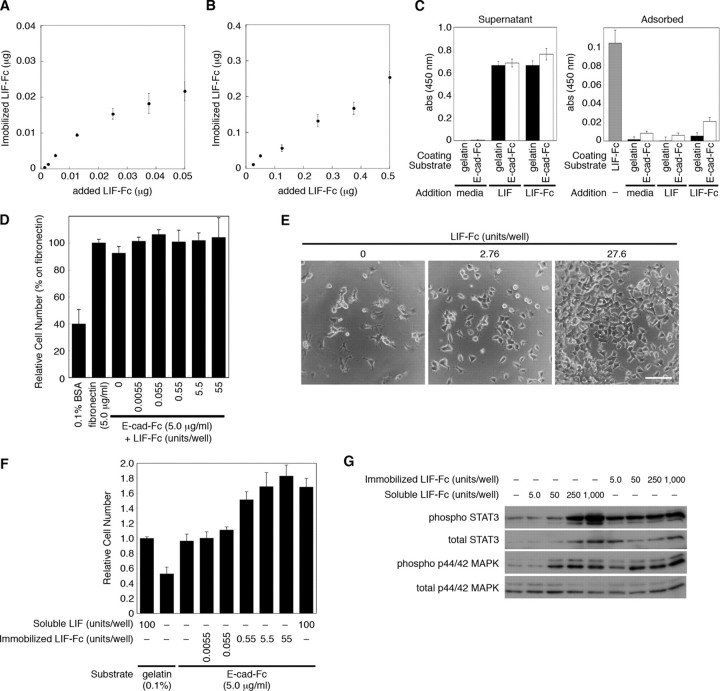
First, as described under “Experimental Procedures,” the amount of LIF-Fc at the co-immobilizing condition was estimated. The amount of adsorbed LIF-Fc reached a plateau at the same concentration of E-cad-Fc (0.25 μg) and gradually increased at the higher concentration than the concentration of E-cad-Fc (Fig. 4, A and B). This indicated that LIF-Fc and E-cad-Fc competitively adsorb to a polystyrene surface. The optimal amount for coating was estimated from these data. Next, we checked the stability of the immobilized LIF-Fc during cell culture. After ES cells were cultured on LIF-Fc- and E-cad-Fc-coated surface for 3 days, the amount of remaining and detached LIF-Fc was analyzed by the ELISA. There were no significant decreases in the amount of immobilized LIF-Fc (Table 2), and LIF-Fc was not detected in the conditioned media of ES cells (data not shown), indicating that LIF-Fc is not incorporated by cells but stably immobilized on a polystyrene surface. Furthermore, there was no significant adsorption of LIF-Fc onto a gelatinized or E-cad-Fc-coated surface (Fig. 4C); therefore, we used a soluble LIF-Fc for the following experiments.
FIGURE 4.
The amount of immobilized LIF-Fc was measured at a low concentration (A) and a high concentration (B) of LIF-Fc co-immobilized with 0.25 μg of E-cad-Fc. The data indicate means ± S.E. of five separate experiments. C, amount of adsorbed LIF-Fc protein onto gelatinized or E-cad-Fc-coated surface was analyzed by ELISA. LIF or LIF-Fc was diluted with the culture media for ES cells at 1,000 units/ml and incubated on the gelatinized or E-cad-Fc-coated surface for 3 days. The amount of adsorbed or noninteracted proteins (Supernatant) was analyzed by ELISA. The surface coated with LIF-Fc (1,000 units/ml) was used as a control to compare the amount of immobilized LIF-Fc. The data represent means ± S.E. of 12 separate experiments. D, adhesion of mouse ES cells onto the surface of 96-well plate coated with 0.1% BSA, 5.0 μg/ml fibronectin, or co-immobilization of 5.0 μg/ml E-cad-Fc and various concentrations of LIF-Fc. After 3 h of incubation ES cells adhered to the co-immobilized surface with equivalent efficiency as to the surface coated with E-cad-Fc alone. The data indicate means ± S.D. of three separate experiments. E and F, ES cells show higher proliferation ability on the co-immobilized surface with E-cad-Fc and LIF-Fc, depending on the dose of LIF-Fc. E, morphological observation of ES cells cultured in the 24-well plate; the surfaces were co-immobilized with E-cad-Fc and various concentrations of LIF-Fc. ES cells scattered each other, and they proliferated dose-dependently. Scale bar indicates 100 μm. F, proliferative activity of ES cells on an indicated surface was evaluated by staining with Alamar Blue reagent. The data indicate means ± S.D. of three separate experiments. G, after culturing for 3 days in a 12-well plate, the activation of STAT3 and MAPK by immobilized LIF-Fc was analyzed by Western blotting.
TABLE 2.
The amount of remaining LIF-Fc after cultivation for 3 days with/without ES cells
The data indicate mean ± S.E. of three separate experiments.
| Amount of LIF-Fc | 1.68 | 4.19 | 8.38 | 12.6 | 16.8 |
|---|---|---|---|---|---|
| 1.68 ng/well | |||||
| % remaining LIF-Fc (without cells) | 137 ± 65 | 121 ± 27 | 104 ± 20 | 93 ± 9.5 | 94 ± 7.7 |
| % remaining LIF-Fc (with cells) | 143 ± 56 | 126 ± 25 | 104 ± 24 | 104 ± 17 | 106 ± 10 |
The adhesion of ES cells onto the co-immobilized surface was not affected by the concentration of LIF-Fc (Fig. 4D), and the cells showed scattering morphology similar to those on the surface coated with E-cad-Fc (Fig. 4E). These results indicate that at the concentration indicated, ES cells stably adhere onto the co-immobilized surface by E-cadherin-mediated adhesion and that a homogeneous environment could be achieved by this system. Furthermore, depending on the concentration of immobilized LIF-Fc, the ES cells showed higher proliferation ability on a co-immobilized surface than on a gelatinized surface (Fig. 4F). The activation of LIF signaling was sustained on the co-immobilized surface even at the low amount of immobilized LIF-Fc (Fig. 4G, 5 units/well). These results also indicate that LIF-Fc is stably immobilized and possesses the ability to induce LIF signaling.
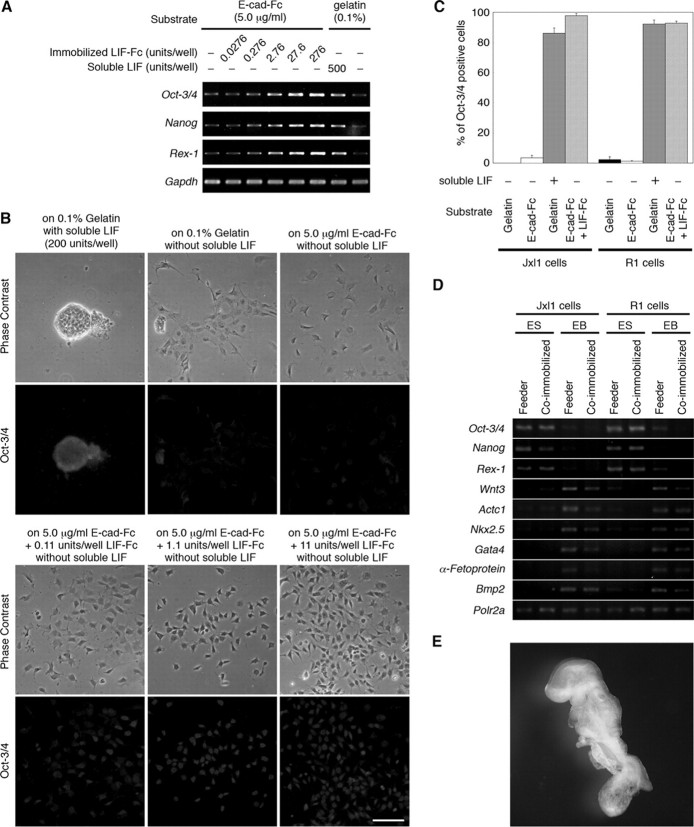
We next examined the ability to maintain undifferentiated phenotypes on a co-immobilized surface. The expression of Oct-3/4 (24), Rex-1 (25), and Nanog (26, 27) genes was maintained in a dose-dependent manner of immobilized LIF-Fc without additional LIF supplementation, and the expression of the genes was retained for at least 15 days. The expression of these genes diminished upon LIF withdrawal (Fig. 5A). Immunostaining also showed that ES cells cultured on co-immobilized dishes with LIF-Fc and E-cad-Fc for 5 days expressed Oct-3/4 in the nucleus, whereas depletion of LIF led to a marked reduction in Oct-3/4 staining (Fig. 5B). These results indicate that a co-immobilized surface of E-cad-Fc and LIF-Fc did effectively maintain the undifferentiated phenotype of ES cells. Furthermore, we tried to apply our culture system to feeder-dependent ES cells. Jxl1 and R1 cells, which are cultured on feeder cells, were seeded on the gelatinized plate or on the co-immobilized surface of E-cad-Fc and LIF-Fc. After culturing for 5 days, the ratio of Oct-3/4-positive cells was calculated. Most of cells cultured on the co-immobilized surface retained the expression of Oct-3/4 (Fig. 5C).
FIGURE 5.
ES cells maintain the undifferentiated phenotype and pluripotency on the co-immobilized surface. A, expression of three genes that are markers of the undifferentiated state was analyzed by RT-PCR. B, ES cells cultured for 5 days in different conditions were stained with anti-Oct-3/4-specific antibody. On the co-immobilized surface, the expression of Oct-3/4 was observed even though low concentration of LIF-Fc was immobilized. Scale bar indicates 100 μm. C, adaptation of feeder-dependent ES cells onto the co-immobilized surface with E-cad-Fc and LIF-Fc. Jxl1 and R1 cells were seeded on the surface coated with several matrices and cultured for 5 days. The ratio of Oct-3/4 positive cells was calculated by ImageJ software. The data indicate mean ± S.E. of five separate experiments. D, ability to differentiate into multilineage cells was assessed by RT-PCR. Feeder-dependent ES cells were seeded on feeder layer (Feeder) or on the co-immobilized surface (Co-immobilized). After being maintained for 10 passages, cells were cultured to form embryoid bodies. Embryoid bodies were cultured for 11 days, and then expression of marker genes was analyzed by RT-PCR. ES, undifferentiated cells; EB, embryoid bodies. Polr2a was used as an internal control. E, feeder-dependent R1 cells were maintained on the co-immobilized surface of E-cad-Fc and LIF-Fc for 10 passages, and then cells were aggregated with four-cell stage tetraploid CD-1 embryos. The embryos were observed at the stage E9.5.
We also checked whether the multidifferentiating potency of ES cells is affected by culturing on a co-immobilized surface. To assess the potential of ES cells to differentiate into multiple cell lineages, embryoid bodies were generated from ES cells that had been cultured for 10 passages on the co-immobilized surface. These embryoid bodies were cultured for an additional 11 days without LIF to induce differentiation. The level of expression of lineage-specific marker genes was analyzed by RT-PCR. Similar to results using the conventional culture methods, embryoid bodies derived from co-immobilized surfaces developed into ectoderm (Wnt3), mesoderm (Actc1 and Nkx2.5), and endoderm (Gata4, α-Fetoprotein, and Bmp2) derivatives (Fig. 5D).
Finally, we confirmed the pluripotent ability of ES cells cultured on co-immobilized surface with E-cad-Fc and LIF-Fc by tetraploid aggregation. ES cells were directly transferred to our system from culture on feeder cells. After 10 passages with one freeze/thaw cycle, ES cells were aggregated with four-cell stage CD-1 tetraploid embryos and transferred to pseudo-pregnant mice. ES cell-derived embryos were recovered at E9.5. These results demonstrate that without LIF supplementation, the coimmobilized surface with LIF-Fc and E-cad-Fc can maintain the undifferentiated state and pluripotency of ES cells, even though the quite different morphology was compared with the conventional culture systems.
DISCUSSION
In this study, we applied two immobilizable model proteins, E-cad-Fc and LIF-Fc, to establish a new culture system of mouse ES cells, and we demonstrated that mouse ES cells can be effectively maintained on a co-immobilized surface with the model proteins. Furthermore, we found that LIF-Fc is not adsorbed onto the E-cad-Fc-coated surface (Fig. 4C) even though LIF-Fc has an Fc region. This result indicates that the surface would be occupied by the initial adsorption of E-cad-Fc, which would prevent the further adsorption of LIF-Fc. Therefore, the preparation of the mixed solution should be important for the co-immobilization of model proteins.
When cultured on the surface coated only with LIF-Fc, mouse ES cells still form aggregated colonies, and the adhesion of cells is weak, even though the cells can maintain undifferentiated phenotypes (Fig. 3). When ES cells form colonies, the signaling from LIF is needed to reach the center of the colonies to maintain an undifferentiated state of ES cells. The soluble LIF might diffuse into the colonies; however, immobilized LIF cannot interact with the inside cells. To explain this issue, we suggest the following hypotheses. 1) LIF-Fc is detached from the surface, and 2) the factors secreted from ES cells by LIF stimulation contribute to maintain the undifferentiated state of ES cells. We analyzed the stability of immobilized LIF-Fc by ELISA and obtained the data that the amount of LIF-Fc did not change after cultivation of ES cells (Table 2). In addition, immobilized LIF-Fc prevented the incorporation of the LIF receptor (Fig. 3E). These results indicate that LIF-Fc was stably immobilized but not incorporated into cells. Furthermore, similar studies suggest that chemically immobilized LIF can be effective for maintenance of murine ES cells (28–30), although the cells also formed aggregated colonies. Because covalent binding is more stable than hydrophobic interaction of LIF-Fc and cannot be digested by enzymatic activity of cells, the results also indicate the possibility of another pathway that transduces the LIF signals into the inside of the colonies. In addition, several studies suggest that colony formation induces the heterogeneous distribution of mouse ES cells (21–23) and also human ES cells (31, 32). Singh et al. (23) reported that the heterogeneous expression of Nanog and SSEA-1 is induced when ES cells are maintained in the colony-forming condition, and that the cells expressing a low level of Nanog express primitive endodermal marker genes. Furthermore, Davey and Zandstra (22) suggested that autocrine non-LIF ligands of gp130 transduce the signaling for maintenance of an undifferentiated state of mouse ES cells, and they also suggested the heterogeneity of autocrine signals. Recently, a number of new culture methods for human ES cells have been developed that do not require feeder cells or serum to reduce the risk of pathogens (33–40). In these culture methods, the undifferentiated human ES cells still form aggregates, and the aggregated colonies are reported to induce heterogeneity of cells (31, 32).
From these findings, we propose that the close contact between cells in these aggregates may lead to paracrine or autocrine interactions that could generate heterogeneity. Therefore, our system could improve the culture methods of mouse ES cells by maintaining them in homogeneous conditions.
We also found that a reduced amount of LIF was required for maintenance of mouse ES cells on a LIF-immobilized surface. Although we did not compare the actual amount of LIF interacting with cells, our results indicate that the immobilization of LIF is more effective in maintaining ES cells than is possible using soluble forms. We previously reported that ES cells maintained as a single cell require less amounts of LIF (11). Therefore, a co-immobilized surface is an efficient system to maintain ES cell properties.
In conclusion, we demonstrated that the culture surface coated with LIF-Fc can be useful to maintain the undifferentiated state of mouse ES cells, and that ES cells cultured without any additional LIF supplementation on the co-immobilized surface with E-cad-Fc and LIF-Fc could be maintained as single cells with complete ES cell features. Moreover, the new culture system can maintain ES cells in the homogeneous culture condition to eliminate the risk of unexpected differentiation, and this system is beneficial for single cell-based assay of signaling pathways and cell responses induced by immobilized growth factors.
Acknowledgments
We thank Prof. Hitoshi Niwa of the RIKEN Center for Developmental Biology and Prof. Andras Nagy of the Samuel Lunenfeld Research Institute, Mount Sinai Hospital for their generous gift of EB3 cells (H.N.) and R1 cells (A.N.).
This work was supported by a grant from the 21st Century Centers of Excellence Program and a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ES, embryonic stem; LIF, leukemia inhibitory factor; FBS, fetal bovine serum; PBS, phosphate-buffered saline; BSA, bovine serum albumin; MAPK, mitogen-activated protein kinase; ELISA, enzyme-linked immunosorbent assay; RT, reverse transcription; E-cad, E-cadherin; ALP, alkaline phosphatase.
References
- 1.Evans, M. J., and Kaufman, M. (1981) Nature 292 154–156 [DOI] [PubMed] [Google Scholar]
- 2.Martin, G. R. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gearing, D. P., Gough, N. M., King, J. A., Hilton, D. J., Nicola, N. A., Simpson, R. J., Nice, E. C., Kelso, A., and Metcalf, D. (1987) EMBO J. 6 3995–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann, H., and Wong, G. G. (1989) J. Immunol. 143 1163–1167 [PubMed] [Google Scholar]
- 5.Ferrara, N., Winer, J., and Henzel, W. J. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 698–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grell, M., Douni, E., Wajant, H., Lohden, M., Clauss, M., Maxeiner, B., Georgopoulos, S., Lesslauer, W., Kollias, G., Pfizenmaier, K., and Scheurich, P. (1995) Cell 83 793–802 [DOI] [PubMed] [Google Scholar]
- 7.Singh, A. B., Tsukada, T., Zent, R., and Harris, R. C. (2004) J. Cell Sci. 117 1365–1379 [DOI] [PubMed] [Google Scholar]
- 8.Tanaka, Y., Kimata, K., Adams, D. H., and Eto, S. (1998) Proc. Assoc. Am. Physicians 110 118–125 [PubMed] [Google Scholar]
- 9.Rathjen, P. D., Toth, S., Willis, A., Heath, J. K., and Smith, A. G. (1990) Cell 62 1105–1114 [DOI] [PubMed] [Google Scholar]
- 10.Mereau, A., Grey, L., Piquet-Pellorce, C., and Heath, J. K. (1993) J. Cell Biol. 122 713–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagaoka, M., Koshimizu, U., Yuasa, S., Hattori, F., Chen, H., Tanaka, T., Okabe, M., Fukuda, K., and Akaike, T. (2006) PLoS ONE 1 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagaoka, M., Ise, H., Harada, I., Koshimizu, U., Maruyama, A., and Akaike, T. (2008) J. Cell. Biochem. 103 296–310 [DOI] [PubMed] [Google Scholar]
- 13.Ogiwara, K., Nagaoka, M., Cho, C. S., and Akaike, T. (2006) Biochem. Biophys. Res. Commun. 345 255–259 [DOI] [PubMed] [Google Scholar]
- 14.Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W., and Roder, J. C. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niwa, H., Masui, S., Chambers, I., Smith, A. G., and Miyazaki, J. (2002) Mol. Cell. Biol. 22 1526–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagaoka, M., and Akaike, T. (2003) Protein Eng. 16 243–245 [DOI] [PubMed] [Google Scholar]
- 17.Metcalf, D., Hilton, D. J., and Nicola, N. A. (1988) Leukemia (Basingstoke) 2 216–221 [PubMed] [Google Scholar]
- 18.Duncan, S. A., Nagy, A., and Chan, W. (1997) Development (Camb.) 124 279–287 [DOI] [PubMed] [Google Scholar]
- 19.Nagy, A., Gocza, E., Diaz, E. M., Prideaux, V. R., Ivanyi, E., Markkula, M., and Rossant, J. (1990) Development (Camb.) 110 815–821 [DOI] [PubMed] [Google Scholar]
- 20.Blanchard, F., Duplomb, L., Wang, Y., Robledo, O., Kinzie, E., Pitard, V., Godard, A., Jacques, Y., and Baumann, H. (2000) J. Biol. Chem. 275 28793–287801 [DOI] [PubMed] [Google Scholar]
- 21.Cui, L., Johkura, K., Yue, F., Ogiwara, N., Okouchi, Y., Asanuma, K., and Sasaki, K. (2004) J. Histochem. Cytochem. 52 1447–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davey, R. E., and Zandstra, P. W. (2006) Stem Cells 24 2538–2548 [DOI] [PubMed] [Google Scholar]
- 23.Singh, A. M., Hamazaki, T., Hankowski, K. E., and Terada, N. (2007) Stem Cells 25 2534–2542 [DOI] [PubMed] [Google Scholar]
- 24.Niwa, H., Miyazaki, J., and Smith, A. G. (2000) Nat. Genet. 24 372–376 [DOI] [PubMed] [Google Scholar]
- 25.Rogers, M. B., Hosler, B. A., and Gudas, L. J. (1991) Development (Camb.) 113 815–824 [DOI] [PubMed] [Google Scholar]
- 26.Mitsui, K., Tokuzawa, Y., Itoh, H., Segawa, K., Murakami, M., Takahashi, K., Maruyama, M., Maeda, M., and Yamanaka, S. (2003) Cell 113 631–642 [DOI] [PubMed] [Google Scholar]
- 27.Chambers, I., Colby, D., Robertson, M., Nichols, J., Lee, S., Tweedie, S., and Smith, A. (2003) Cell 113 643–655 [DOI] [PubMed] [Google Scholar]
- 28.Makino, H., Hasuda, H., and Ito, Y. (2004) J. Biosci. Bioeng. 98 374–379 [DOI] [PubMed] [Google Scholar]
- 29.Cetinkaya, G., Turkoglu, H., Arat, S., Odaman, H., Onur, M. A., Gumusderelioglu, M., and Tumer, A. (2007) J. Biomed. Mater. Res. 81 911–919 [DOI] [PubMed] [Google Scholar]
- 30.Alberti, K., Davey, R. E., Onishi, K., George, S., Salchert, K., Seib, F. P., Bornhäuser, M., Pompe, T., Nagy, A., Werner, C., and Zandstra, P. W. (2008) Nat. Meth. 5 645–650 [DOI] [PubMed] [Google Scholar]
- 31.Stewart, M. H., Bosse, M., Chadwick, K., Menendez, P., Bendall, S. C., and Bhatia, M. (2006) Nat. Meth. 3 807–815 [DOI] [PubMed] [Google Scholar]
- 32.Bendall, S. C., Stewart, M. H., Menendez, P., George, D., Vijayaragavan, K., Werbowetski-Ogilvie, T., Ramos-Mejia, V., Rouleau, A., Yang, J., Bosse, M., Lajoie, G., and Bhatia, M. (2007) Nature 448 1015–1021 [DOI] [PubMed] [Google Scholar]
- 33.Xu, C., Inokuma, M. S., Denham, J., Golds, K., Kundu, P., Gold, J. D., and Carpenter, M. K. (2001) Nat. Biotechnol. 19 971–974 [DOI] [PubMed] [Google Scholar]
- 34.Amit, M., Shariki, C., Margulets, V., and Itskovitz-Eldor, J. (2004) Biol. Reprod. 70 837–845 [DOI] [PubMed] [Google Scholar]
- 35.Ying, Q. L., Nichols, J., Chambers, I., and Smith, A. (2003) Cell 115 281–292 [DOI] [PubMed] [Google Scholar]
- 36.Qi, X., Li, T. G., Hao, J., Hu, J., Wang, J., Simmons, H., Miura, S., Mishina, Y., and Zhao, G. Q. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa, K., Matsui, H., Ohtsuka, S., and Niwa, H. (2004) Genes Cells 9 471–477 [DOI] [PubMed] [Google Scholar]
- 38.Sato, N., Meijer, L., Skaltsounis, L., Greengard, P., and Brivanlou, A. H. (2004) Nat. Med. 10 55–63 [DOI] [PubMed] [Google Scholar]
- 39.James, D., Levine, A. J., Besser, D., and Hemmati-Brivanlou, A. (2005) Development (Camb.) 132 1273–1282 [DOI] [PubMed] [Google Scholar]
- 40.Beattie, G. M., Lopez, A., Bucay, N., Hinton, A., Firpo, M. T., King, C. C., and Hayek, A. (2005) Stem Cells 23 489–495 [DOI] [PubMed] [Google Scholar]