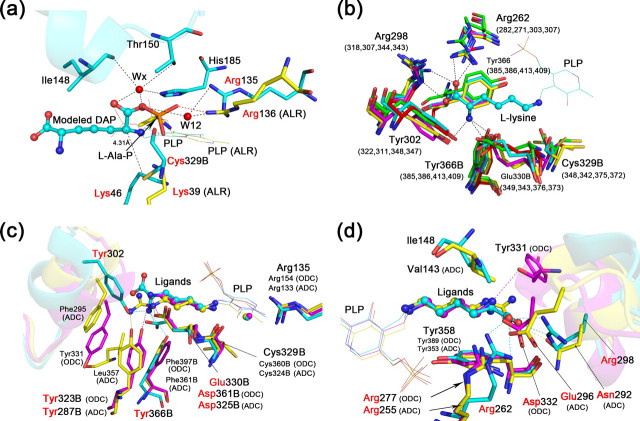
FIGURE 3.
Comparison of the substrate-binding sites among group IV PLP-dependent decarboxylase. a, modeled HpDAPDC-DAP structure (cyan) is superposed with the BsALR-l-Ala-P structure (yellow). The active site loop of HpDAPDC is drawn as ribbon. Residues with red names are of particular interest and described in detail in the text (same below). b, substrate-binding residues of various DAPDCs are superposed. Only the residues of HpDAPDC are named. The numbers in parentheses indicate corresponding residues from other species. c and d, superposition of HpDAPDC (cyan), TbODC (magenta), and cvADC (yellow). The two figures are viewed toward the l-stereocenter amine (c) and carboxyl (d) of the bound l-lysine in HpDADPC, respectively. The specificity element helices are shown as ribbon.