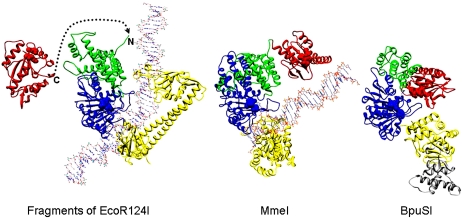
Figure 7.
Structural evolution of type IIG RM enzymes from a type I RM enzyme undergoing fusion of the C terminus of an endonuclease domain from HsdR, via deletion of the motor domains, to the N terminus of HsdM. The structure on the left shows part of EcoR124I, with one endonuclease domain from HsdR (in red), one HsdM (N-terminal domain is in green, and the MTase catalytic domain is in blue), and the HsdS (in yellow) (two TRDs). DNA bound to the MTase core is shown, but DNA bound to HsdR is omitted for clarity. The dashed line shows how the end of the endonuclease domain could join with the N terminus of HsdM to form a structure similar to the type IIG structures shown on the right. The catalytic motifs in the endonuclease domain and HsdM are shown in spacefill. The middle structure shows the structural model of MmeI with bound DNA with the same coloring used for equivalent domains (endonuclease domain, N-terminal domain, MTase catalytic domain, and TRD) (Nakonieczna et al. 2009; coordinates from ftp://genesilico.pl/iamb/models/RM.MmeI). The structure on the right shows the crystallographic structure of BpuSI (PDB code: 3s1s) with the same coloring of domains as in the other structures and with an inserted extra domain shown in gray (Shen et al. 2011). DNA is absent in this structure, and one can see that the endonuclease domain would be blocking the DNA-binding site on the TRD. Shen et al. (2011) proposed that the endonuclease domain would twist away to allow DNA sequence recognition.