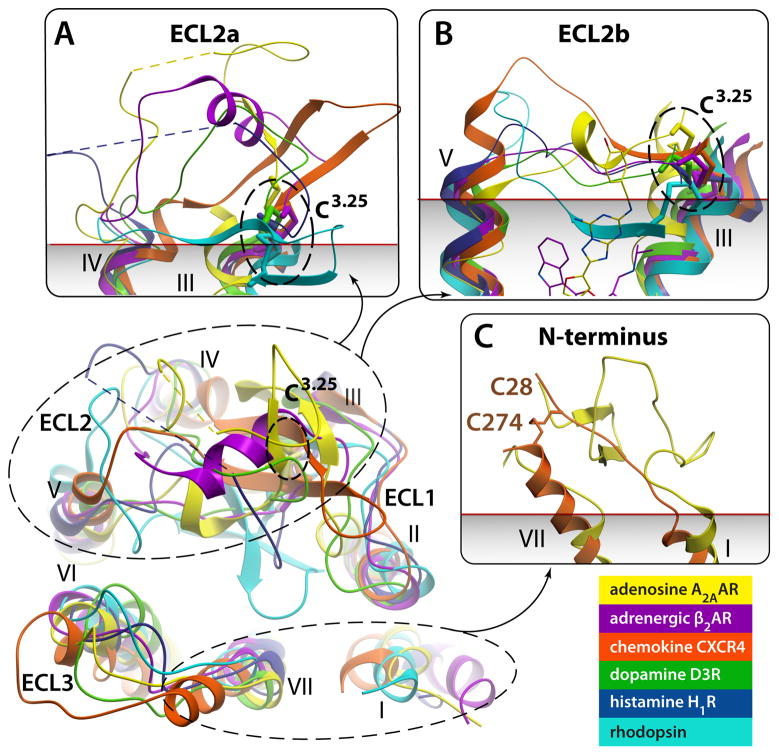
Fig. 3.
Remarkable diversity in the EC region. Top view of ECLs from superimposed GPCR structures with close-up side views in panels (A)–(C). Note different secondary structure elements (or lack thereof) in ECL2, as well as large deviations in positions of extracellular tips of the TM helices. N-termini of Rhodopsin and CXCR4 are removed for clarity. (A) Close up of ECL2a, a part of ECL2 from helix IV to the conserved disulfide bond connecting ECL2 to helix III. The ECL2a is highly variable in length and secondary structure elements, showing distinct structure between structures of all GPCRs solved; the only exception being the practically identical ECL2a conformations ((RMSD(Cα)=0.5 A)) between very closely related β2AR (PDB ID 2RH1) and β1AR (PDB ID 2VT4, not shown). Short unresolved portions of ECL2a in A2AAR and H1R are shown by dashed lines. (B) Close up side view of ECL2b – the linker part connecting helices V and III through the disulfide bond to Cys3.25. Note that in some GPCRs the ECL2b element is as short as 4–5 amino acids and fully extended, while longer sequence in others can comprise small secondary structure elements. Relative positions of Carazolol and ZM241385 interacting with ECL2b residues are shown by thin lines. (C) N-terminus is fully resolved in Rhodopsin (PDB ID 1GZM; residues 1:34) and partially in CXCR4 (PDB ID 3ODU; residues 27:34) structures; in other known structures N-termini are likely disordered. Approximate membrane boundary is shown in panels (A)–(C), as predicted by OPM database [86].