Abstract
Nucleophosmin (NPM) is a multifunctional nucleolar protein that has been linked with nucleolar stress. In non-neuronal cell lines, NPM may enhance or inhibit the activity of tumor suppressor p53, a major apoptotic protein. The relationship between NPM and p53 in the central nervous system (CNS) remains unknown. Here, we assessed the role of NPM in the CNS using a model of seizure-induced neurodegeneration. We show that NPM overexpression is neuroprotective against kainic acid-induced excitotoxicity, and that downregulation of NPM is pro-apoptotic in a p53-independent manner. These results suggest a key role for NPM in promoting neuronal survival and a novel mechanism of neuronal degeneration triggered by nucleolar stress.
Keywords: Nucleophosmin, nucleolar stress, tumor suppressor p53, neuronal degeneration, kainic acid, siRNA
1. Introduction
The nucleolus, a non-membrane bound intranuclear structure, is well-known for its role in ribosomal RNA synthesis, ribosome assembly and diverse cellular processes relevant to cell growth and proliferation, cell cycle control and tumorgenesis [1]. Recent evidence has also implicated the nucleolus in the cellular response to stress [1,2]. In this regard, UV radiation, hypoxia and other cytotoxic agents, may result in nucleolar stress characterized by disruption of nucleolar structure, redistribution of nucleolar proteins, and inhibition of nucleolar function [1,3,4].
Nucleophosmin (NPM) is a multifunctional protein that has been shown to play an important role in the nucleolar response to stress [1,2]. NPM levels are closely correlated with mitogen-induced cell proliferation, and NPM fusion proteins are found in several malignancies [3,4]. Importantly, NPM directly participates in ribosome assembly [5], chromatin remodeling and centrosome duplication [5,6]. During nucleolar stress NPM may stabilize tumor suppressor p53, a major regulator of apoptosis, or promote its degradation [1,7]. The relationship between NPM and p53 varies with cell type and/or stimulus [7,8], Thus, NPM may block apoptosis by preventing p53 mitochondrial translocation, but can also interact with the pro-apoptotic protein, Bax, and promote cell death [9,10]. In contrast to non-neural systems, the role of NPM in the CNS is poorly understood. Here we demonstrate a link between NPM abundance and neuronal survival following excitotoxic injury. Studies in cultured neurons show that NPM downregulation is pro-apoptotic in a p53-independent manner. These results suggest a key role for the nucleolar protein, NPM, as a promoter of neuronal survival.
2. Materials and methods
2.1. Animals and drug treatment
Sprague Dawley rats (4 weeks old) obtained from Charles River were injected subcutaneously with the excitotoxin, kainic acid (KA, 10 mg/kg) (N=5-7) or vehicle control (10 mM Tris-HCl) (N=5) [11]. Following KA administration animals were observed for at least 4 hr for spontaneous seizures. Only those animals exhibiting status epilepticus were included in the study. Rats were sacrificed under CO2 gas anesthesia at indicated time points after KA administration and the brain was rapidly removed and dissected [11]. Breeding pairs of p53-deficient mice on an FVB background obtained from The Jackson Laboratory (Bar Harbor, ME) were bred by crossing p53+/− with either p53+/− or p53−/− mice and confirmed by genotyping. Wild type (p53+/+) littermates were used as controls. Mice 4 weeks postnatal were given KA (20 mg/KA, s.c.) and processed similar to rats. All animals were treated in accordance with the guidelines of the UC Irvine School of Medicine and housed at the vivarium on a 12 hr light/dark cycle with food and water available ad libitum.
2.2. Cell culture and treatments
Primary cortical neuron cultures were prepared from embryonic day 18 (E18) Sprague-Dawley fetuses and cultured in neurobasal media as described [11]. Human SH-SY5Y neuroblastoma cell line and NE-4C cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). SH-SY5Y cells were cultured with a 1:1 mixture of DMEM and F12 media, 10% FBS, 0.2% PenStrep and 5 mM glutamine. NE-4C neuroectodermal cells were cultured in DMEM, 10% FBS, 0.2% PenStrep and 5 mM glutamine. Primary neurons or SH-SY5Y cells were treated with KA (100-400 μM) for 10 min [11]. For CPT treatments, 0.05-1.0 μM was directly added to fresh culture media. At indicated times following treatments cells were either fixed in 90% cold methanol or harvested for isolation of total RNA or preparation of whole cell lysates [11].
2.3. Constructs and transfections
Plasmids containing Flag-tagged wild type p53, mutant p53R175Q, dsp53TAF, and a Flag-empty vector were constructed from Clontech plasmids [12]. Mixtures containing Fugene 6 and either 2 μg of total plasmid DNA or 150 nM siRNA (control or NPM targeted) were incubated with cells as per the manufacturer’s manual. For NPM overexpression assays, either Flag-NPM or a Flag-empty vector was transfected into SH-SY5Y cells on chamber slides followed by KA treatment (100 μM KA for 10 min) 16 hr later. Cells were fixed with 90% cold methanol and processed for dual labeling of TUNEL and Flag [11,12].
2.4. Immunohistochemistry and TUNEL staining
Immunohistochemistry was performed as described [11] using the following dilutions of primary antibody: anti-NPM,1:800; p53 CM5,1:200; Mdm2 SMP14, 1:100; anti-p53 full length, 1:450; anti-Bax 1:200; anti-cytochrome C, 1:500; and anti-caspase 3 CM1, 1:300. Secondary antibody incubations were with biotin- (1:200 dilution), Cy3- (1:80 dilution), or FITC- (1:110 dilution) conjugated antibodies for 30 min. Confocal microscopy was performed using a Zeiss LSM510-META microscope. For terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), samples were labeled with an In Situ cell death detection kit [12].
2.5. Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted using Trizol reagent and RT-PCR was performed using three different pairs of primers to confirm specificity [11]. Primers used for all of the RT-PCR reactions are listed in Table 1 (see Supplementary data). Amplification of total RNA without the RT reaction served as a negative control.
2.6. Western blotting
Briefly, 10 or 20 μg total protein lysate for each sample was resolved by 10% SDS-PAGE, transferred onto an immobilon-P membrane, blocked with Blotto solution, and incubated with primary antibody [1:100 (NPM N-terminal, p53, Mdm2), 1:1000 (NPM c-terminal, β-actin)] overnight at 4°C followed by incubation with an appropriate HRP-conjugated secondary antibody. Immunoreactivity was visualized by ECL [12].
2.7. Quantification and statistical analysis
For NPM or NPM/TUNEL co-localization, a total of 1000 cells in brain sections (three sections/group) or 100 cultured cells were counted randomly in 10 different fields. In the Flag-NPM overexpression assays, TUNEL-positive cells were counted based on the total number of 100 Flag-positive cells on each slide transfected with either Flag-NPM or Flag-empty vector constructs. Quantification of gel pixel intensity was performed with Scientific Imaging System KODAK ID software. The data are depicted as the mean ± S.E.M. from 3 to 5 independent experiments. Statistical analysis was performed by one-way ANOVA followed by Student’s t-test. The differences between groups were considered statistically significant when p ≤ 0.05.
3. Results
3.1. Downregulation of NPM abundance following prolonged seizures
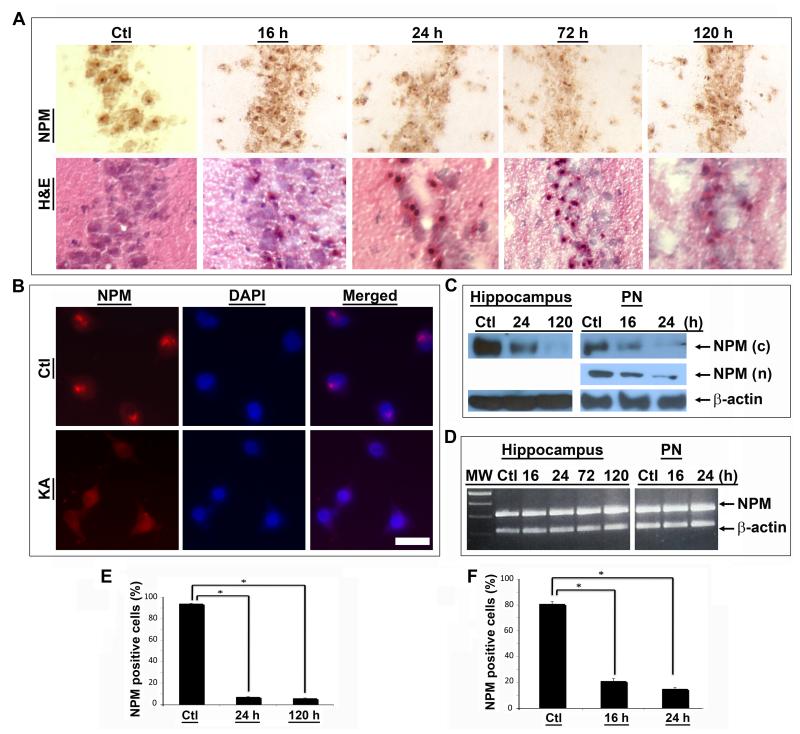
Systemic administration of KA, a potent glutamate analogue, induces prolonged seizures, selective neuronal degeneration and up regulation of p53 and related proteins linked with DNA damage in degenerating neurons [13,14]. KA-mediated excitotoxicity was therefore employed to explore the relationship between NPM, nucleolar stress and neuronal degeneration. As shown in Figure 1, there was prominent NPM immunoreactivity in the nucleolus of hippocampal CA1 neurons from vehicle-treated rats. In contrast, nucleolar NPM immunoreactivity was significantly reduced after KA treatment (Fig. 1A, top row). Importantly, adjacent brain sections stained with hematoxylin and eosin (H&E) exhibited eosinophilic cells with pyknotic nuclei, i.e., morphological signs of irreversible cell damage, in the same regions displaying decreased NPM levels (Fig. 1A, bottom row). To corroborate these findings, loss of nucleolar NPM immunoreactivity was observed in primary neurons treated with KA (Fig. 1B). Quantification of the results showed that a significant decrease in NPM occurred in both the hippocampus and primary neurons after KA treatment (Fig. 1E&F). These observations were confirmed by immunoblot analyses using antibodies against the C- or N-terminus of NPM. The results showed decreased NPM abundance in both the hippocampus and primary neurons after KA treatment (Fig. 1C).
Figure 1. Downregulation of NPM following KA-induced excitotoxicity.
(A) Immunohistochemistry using a C-terminal NPM antibody reveals a significant decrease in nucleolar NPM staining in the CA1 region of the hippocampus of rats treated with KA compared with an untreated control rat (Ctl) (top row). H&E staining of adjacent sections demonstrates normal morphology in a control rat and neuronal damage at indicated times following KA treatment (bottom row). (B) Immunofluorescence microscopy demonstrates strong nucleolar staining of NPM in untreated primary neurons (Ctl) and decreased NPM immunoreactivity in neurons 24 hr after KA treatment (KA). Nuclei stained with DAPI. Scale bar = 20 μm. (C) Western blotting using the same C-terminal NPM antibody (c) reveals a reduction of NPM protein abundance in rat hippocampus following onset of KA-induced seizures and in primary neurons (PN) after KA treatment relative to untreated controls (Ctl). Similar changes were observed in primary neurons using an NPM N-terminal antibody (n). β-actin immunoreactivity is shown as internal loading control. (D) RT-PCR analysis demonstrates no significant changes in levels of NPM mRNA in the hippocampus of control (Ctl) and KA-treated rats or primary neurons (PN) at indicated times after treatment. β-actin product is used as a loading control. (E, F) Quantification of NPM immunoreactive cells in the hippocampal CA1 region (e) and primary neurons after KA treatment (F) demonstrates a significant decrease in the number of NPM(+) cells at indicated times after KA treatment in comparison with controls.
To determine if changes in NPM gene transcription paralleled those in NPM protein, mRNA was isolated from both whole hippocampi or cultured neurons and nested RT-PCR performed using different amplification cycles and two sets of primers for the flanking and internal regions of the NPM gene. There was no significant change in the level of NPM mRNA up to 120 hr after KA treatment (Fig. 1D). As shown in the same figure, KA did not affect NPM mRNA levels in rat primary neurons.
3.2. NPM downregulation occurs in degenerating neurons
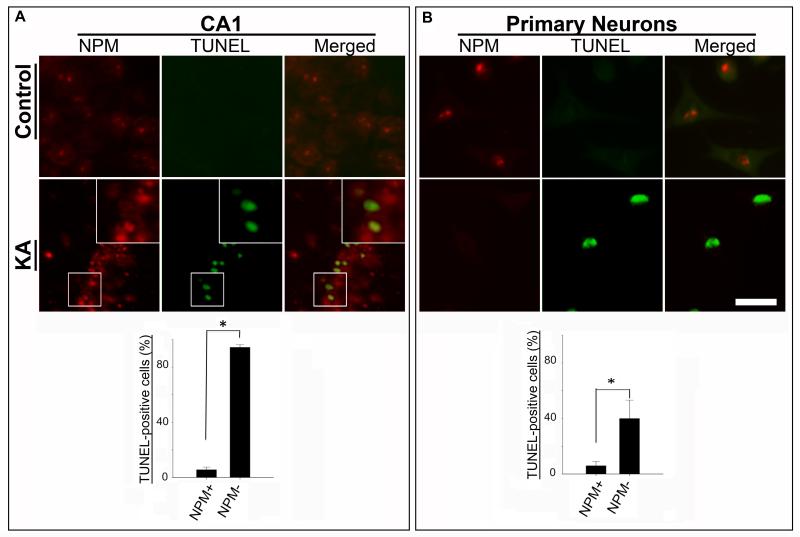
To further examine the relationship between NPM and neuronal degeneration, brain sections were dual-labeled for NPM and TUNEL. The results revealed decreased NPM expression in TUNEL-positive neurons after KA treatment (Fig. 2A). In comparison, there was abundant nucleolar NPM immunoreactivity in TUNEL-negative neurons from control animals. An identical inverse relationship between NPM and TUNEL was observed in primary neurons (Fig. 2B), and in both cases the results were statistically significant (Figs. 2A & B).
Figure 2. NPM is reduced in nucleoli of damaged neurons.
Double-label immunofluorescence shows that nucleoli of cells in the hippocampal CA1 region from untreated control rat or primary neurons are NPM(+) (red) and TUNEL(−). Following KA-treatment the majority of cells in either CA1 or in culture exhibit reduced levels of nucleolar NPM immunoreactivity and are TUNEL(+) (green). Quantification of TUNEL(+) cells (%) that are either NPM(+) or NPM(−) for each treatment group is depicted as mean ± SEM; * denotes p < 0.05.
3.3. Downregulation of NPM leads to increased p53 abundance and neuronal degeneration
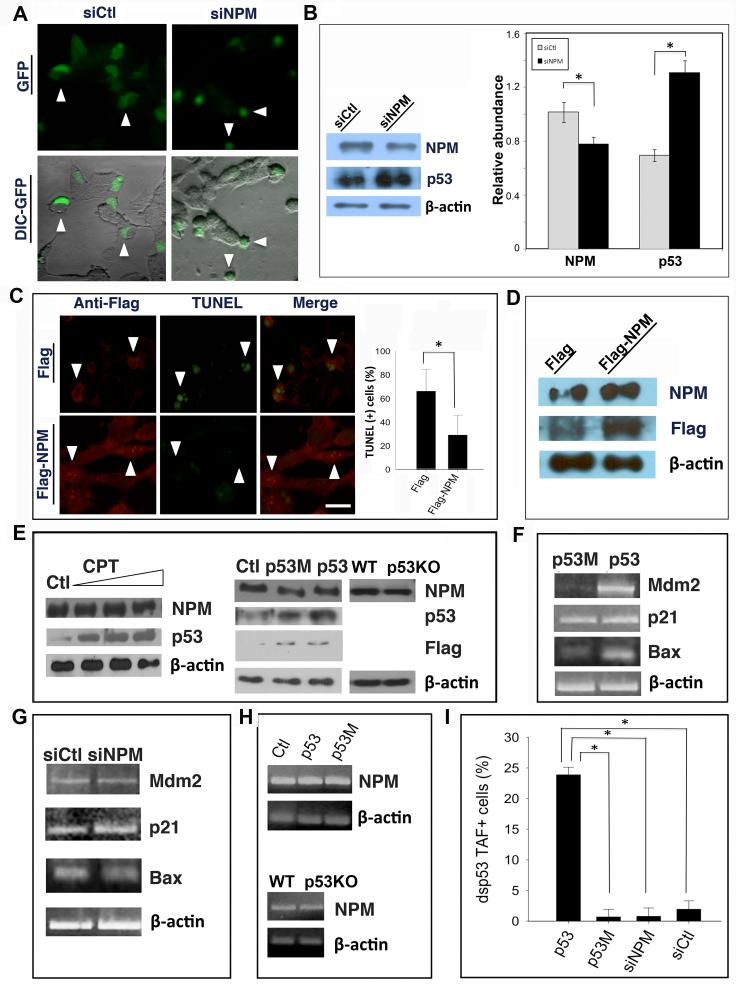
To evaluate whether NPM promotes neuronal survival, the effects of reducing NPM levels were investigated in SH-SY5Y human neuroblastoma cells. Following co-transfection of NPM siRNA and a shuttle plasmid expressing the green fluorescent protein (GFP) reporter, morphological analysis by differential interference contrast (DIC) microscopy revealed cell body shrinkage and membrane blebbing, which are features of apoptosis (Fig. 3A). Western blot analysis showed a 25% reduction of NPM protein 16 hr after NPM siRNA transfection (Fig. 3B). Notably, siRNA-mediated downregulation of NPM resulted in an increase in p53 abundance, suggesting that there may be a regulatory relationship between NPM and p53 in neurons.
Figure 3. NPM overexpression promotes neuronal survival in a p53-independent manner.
(A) SH-SY5Y cells were co-transfected with pEGFP plus either NPM siRNA (siNPM) or a nonsense control siRNA (siCtl), and morphology visualized by fluorescence/DIC microscopy. GFP signal (green) indicates transfected cells, which exhibit normal morphology in the control group (arrowheads) and a shrunken appearance after NPM siRNA transfection (arrowheads). (B) Western blotting using NPM C-terminal antibody with quantification confirms a significant downregulation of NPM and induction of p53, relative to β-actin control, after transfection with NPM siRNA. (C) SH-SY5Y cells were transfected with plasmids expressing either Flag-NPM or Flag alone and then treated with KA. Dual fluorescence labeling using an anti-Flag antibody reveals that cells transfected with Flag-NPM are TUNEL(−) (arrowheads, lower panels), whereas most cells transfected with Flag alone are TUNEL+ (arrowheads, upper panels) by 24 hr after KA treatment. Quantification shows a significant reduction in the number of TUNEL(+) cells following transfection with Flag-NPM. (D) Western blot with either NPM or Flag antibodies confirms overexpression of NPM or Flag following Flag-NPM transfection in SH-SY5Y cells. β-actin was used as a loading control. (E) Left panels: SH-SY5Y cells were treated with increasing concentrations of camptothecin (CPT, 0.05~1.0 μM). Western blotting shows that there is no effect of CPT on NPM abundance in contrast to increased levels of endogenous p53. Right panels: SH-SY5Y cells were transfected with a plasmid expressing either Flag (Ctl), Flag-p53 (p53) or Flag-p53R173Q (p53M). Western blotting using a panel of antibodies as indicated shows no difference in NPM abundance in control cells compared with cells overexpressing either Flag-tagged wild type or mutant p53. NPM abundance is also unchanged in hippocampal lysates from p53 knockout mice (p53KO) compared with wild type (WT) controls. β-actin was used as a loading control. (F) RT-PCR analyses of p53-regulated genes Mdm2, p21 and Bax in SH-SY5Y cells after overexpression of either p53 or p53M or (G) following siRNA transfection. (H) Upper: RT-PCR analysis of NPM expression in SH-SY5Y cells transfected with either p53 or p53M plasmids. Lower: RT-PCR analysis of NPM expression in hippocampus of either p53KO or wild type mice. β-actin was used as an internal loading control. (I) SH-SY5Y cells were co-transfected with dsp53TAF, a reporter of p53 transcriptional activity, and either p53-GFP, p53R173Q-GFP (p53M), siNPM or siCtl plus pEGFP. Bar graphs show the number of p53TAF+ cells (%) following transfection with each of the indicated reagents. Bar graphs in (B), (C) and (I) are depicted as mean ± SEM; * denotes p < 0.05. Scale bar = 10 μm in (A) and (C).
3.4. Overexpression of NPM is neuroprotective
As NPM abundance was significantly reduced following neuronal injury, we explored the possibility that NPM could be neuroprotective. SH-SY5Y cells transfected with either a Flag-tagged NPM or control Flag vector were treated with KA. Dual immunostaining revealed that the number of TUNEL-positive cells was reduced in samples transfected with Flag-NPM (Fig. 3C). Western blotting confirmed that there was a significant increase in NPM abundance following Flag-NPM transfection (Fig. 3D).
3.5. Lack of a regulatory relationship between NPM and p53
To explore the possibility of a regulatory relationship between p53 and NPM in neurons, SH-SY5Y cells were transfected with expression vectors encoding either Flag-tagged p53, p53R175H, a mutated form of p53 that is deficient in DNA binding activity [15] or Flag alone as a control. Western blotting demonstrated an increase in p53 abundance (Fig. 3E, middle panels) and up-regulation of p53-regulated genes Mdm2, p21 and Bax, after transfection of Flag-p53, but not Flag-p53R175H or Flag alone (Fig. 3F). p53 transfection also had no effect on the abundance of either NPM protein (Fig. 3E, middle panels) or mRNA (Fig. 3H, upper panels). NPM mRNA and protein levels were similarly unchanged in the brain of both untreated and KA-treated p53-deficient mice, in which there was minimal neuronal degeneration (Fig. 3E, middle panels and 3H, lower panels; Fig. 5 in Supplementary data)[16]. These results indicated that neither upregulation nor absence of p53 affected NPM expression. Further, RNAi-mediated downregulation of NPM, which resulted in increased p53 abundance (Fig. 3B), had no effect on the expression of p53-regulated genes (Fig. 3G). This was corroborated with a reporter of p53 transcriptional activity (dsp53TAF) (Fig. 3I). We next examined the effects of a different type of neuronal injury on NPM and p53 expression. Cultured neurons were treated with the DNA topoisomerase I inhibitor, camptothecin (CPT), which triggers p53-mediated neuronal apoptosis [11]. While CPT-treated cells exhibited a dose-dependent induction of p53, no significant changes in NPM abundance were detected (Fig. 3E, left panels).
3.6. NPM downregulation leads to p53-independent apoptosis
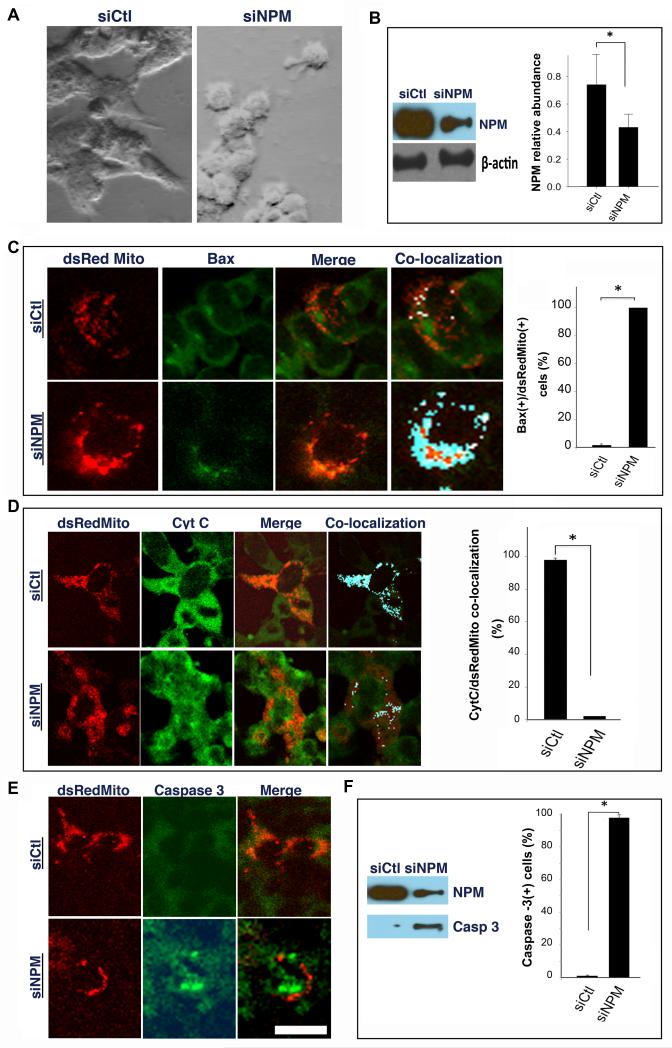
The absence of p53 transcriptional activation suggested that p53 was not required for NPM siRNA-mediated neuronal degeneration. To explore this further, we studied the effects of downregulating NPM in NE-4C cells, a neuroectodermal cell line that lacks functional p53 [17]. NE-4C cells transfected with NPM siRNA demonstrated morphological changes consistent with apoptosis (Fig. 4A) and a significant reduction in NPM abundance by 16 hr (Fig. 4B). NE-4C cells co-transfected with siNPM plus the mitochondrial reporter, dsRedMito, also exhibited a significant increase in mitochondrial Bax translocation (Fig. 4C), cytochrome C release from mitochondria (Fig. 4D) and activation of caspase-3 (Fig. 4E), confirmed by immunoblotting (Fig. 4F).
Figure 4. Downregulation of NPM induces cell death in p53-deficient cells.
(A) Photomicrographs demonstrate normal morphology of NE-4C cells transfected with control siRNA (left) and shrunken appearance 12 hr following transfection of NPM siRNA (right). (B) NPM level is reduced relative to β-actin 16 hr after transfection with NPM siRNA. (C-E) NE-4C cells were co-transfected with dsRedMito, a mitochondrial reporter, and either control (upper rows) or NPM siRNA (lower rows) followed by immunostaining for Bax, cytochrome C or activated caspase-3. Confocal microscopy shows increased (C) Bax mitochondrial co-localization; (D) mitochondrial cytochrome c release and (E) activated caspase-3 following siRNA-mediated downregulation of NPM. (F) Immunoblot shows NPM and activated caspase-3 expression after siRNA treatment. (C, D, F) Bar graphs corresponding to each marker show a significant increase following siNPM transfection with the results depicted as mean ± SEM. * denotes p < 0.05. Scale bar = 20 μm.
4. Discussion
Recent evidence indicates that the nucleolus is a sensor of cell stress and effector of apoptotic cell death [2]. While the mechanisms leading to nucleolar stress and subsequent cell death are not well understood, evidence indicates that NPM plays an important role [1]. The results shown here support the idea that downregulation of NPM is a component of the nucleolar stress response leading to neuronal apoptosis. These results extend current knowledge about the role of NPM to excitotoxic cell death in the CNS. Further, the decrease in NPM abundance in injured neurons likely occurs post-transcriptionally. The significance of NPM in the CNS is underscored by the demonstration that maintenance of NPM expression following injury is neuroprotective.
Despite the connection between NPM expression and neuronal viability, there was no clear association between NPM and p53. Thus, (1) siRNA-mediated downregulation of NPM was associated with accumulation of a transcriptionally-inactive form of p53, (2) NPM levels were unaffected by either p53 overexpression, CPT treatment or absence of p53 in mice and (3) NPM downregulation triggered apoptosis in the absence of p53. While we could not demonstrate a regulatory relationship between NPM and p53 in neurons, the possibility remains that p53 activation may occur upstream from changes in NPM under excitotoxic conditions. .
In summary, our findings indicate that the multifunctional nucleolar protein, NPM, which is essential for early development, may also be critical for maintaining neuronal viability in the injured CNS. Importantly, the results suggest that downregulation of NPM may be an early component of a nucleolar stress response and p53-independent apoptosis. In addition to NPM, the nucleolar response to cellular stress may involve other nucleolar proteins that react to pro-apoptotic signals and could provide novel targets for neuroprotective strategies.
Supplementary Material
Highlights.
Kainic acid-induced excitotoxicity downregulates nucleophosmin in the central nervous system;
Downregulation of nucleophosmin is associated with neuronal cell death in a p53-independent manner;
Restoration of nucleophosmin level in the central nervous system is neuroprotective.
Acknowledgements
The study was supported by NIH AG26637 to Z.T., NS41433 to S.S.S. and the UCMEXUS-CONACYT International Scholarship Program for E.M.M.L..
Abbreviations
- CNS
central nervous system
- DIC
differential interference contrast
- GFP
green fluorescent protein
- NPM
nucleophosmin
- KA
kainic acid
- siRNA
small interfering RNA
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- PCR
polymerase chain reaction
- RT
reverse transcription
- UV
ultraviolet light
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
References
- [1].Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- [2].Mayer C, Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2005;4:1036–1038. doi: 10.4161/cc.4.8.1925. [DOI] [PubMed] [Google Scholar]
- [3].Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- [4].Wasik MA, Zhang Q, Marzec M, Kasprzycka M, Wang HY, Liu X. Anaplastic lymphoma kinase (ALK)-induced malignancies: novel mechanisms of cell transformation and potential therapeutic approaches. Semin Oncol. 2009;36:S27–35. doi: 10.1053/j.seminoncol.2009.02.007. [DOI] [PubMed] [Google Scholar]
- [5].Huang N, Negi S, Szebeni A, Olson MO. Protein NPM3 interacts with the multifunctional nucleolar protein B23/nucleophosmin and inhibits ribosome biogenesis. J Biol Chem. 2005;280:5496–5502. doi: 10.1074/jbc.M407856200. [DOI] [PubMed] [Google Scholar]
- [6].Frehlick LJ, Eirin-Lopez JM, Ausio J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29:49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- [7].Li J, Zhang X, Sejas DP, Pang Q. Negative regulation of p53 by nucleophosmin antagonizes stress-induced apoptosis in human normal and malignant hematopoietic cells. Leuk Res. 2005;29:1415–1423. doi: 10.1016/j.leukres.2005.05.005. [DOI] [PubMed] [Google Scholar]
- [8].Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dhar SK, St Clair DK. Nucleophosmin blocks mitochondrial localization of p53 and apoptosis. J Biol Chem. 2009;284:16409–16418. doi: 10.1074/jbc.M109.005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kerr LE, Birse-Archbold JL, Short DM, McGregor AL, Heron I, Macdonald DC, Thompson J, Carlson GJ, Kelly JS, McCulloch J, Sharkey J. Nucleophosmin is a novel Bax chaperone that regulates apoptotic cell death. Oncogene. 2007;26:2554–2562. doi: 10.1038/sj.onc.1210044. [DOI] [PubMed] [Google Scholar]
- [11].Tan Z, Tu W, Schreiber SS. Downregulation of free ubiquitin: a novel mechanism of p53 stabilization and neuronal cell death. Brain Res Mol Brain Res. 2001;91:179–188. doi: 10.1016/s0169-328x(01)00117-6. [DOI] [PubMed] [Google Scholar]
- [12].Tan Z, Sun X, Hou FS, Oh HW, Hilgenberg LG, Hol EM, van Leeuwen FW, Smith MA, O’Dowd DK, Schreiber SS. Mutant ubiquitin found in Alzheimer’s disease causes neuritic beading of mitochondria in association with neuronal degeneration. Cell Death Differ. 2007;14:1721–1732. doi: 10.1038/sj.cdd.4402180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sakhi S, Bruce A, Sun N, Tocco G, Baudry M, Schreiber SS. p53 induction is associated with neuronal damage in the central nervous system. Proc Natl Acad Sci U S A. 1994;91:7525–7529. doi: 10.1073/pnas.91.16.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tan Z, Sankar R, Tu W, Shin D, Liu H, Wasterlain CG, Schreiber SS. Immunohistochemical study of p53-associated proteins in rat brain following lithium-pilocarpine status epilepticus. Brain Res. 2002;929:129–138. doi: 10.1016/s0006-8993(01)03360-1. [DOI] [PubMed] [Google Scholar]
- [15].Cao Y, Gao Q, Wazer DE, Band V. Abrogation of wild-type p53-mediated transactivation is insufficient for mutant p53-induced immortalization of normal human mammary epithelial cells. Cancer research. 1997;57:5584–5589. [PubMed] [Google Scholar]
- [16].Morrison RS, Wenzel HJ, Kinoshita Y, Robbins CA, Donehower LA, Schwartzkroin PA. Loss of the p53 tumor suppressor gene protects neurons from kainate-induced cell death. J Neurosci. 1996;16:1337–1345. doi: 10.1523/JNEUROSCI.16-04-01337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schlett K, Herberth B, Madarasz E. In vitro pattern formation during neurogenesis in neuroectodermal progenitor cells immortalized by p53-deficiency. Int J Dev Neurosci. 1997;15:795–804. doi: 10.1016/s0736-5748(97)00015-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.