FIGURE 1.
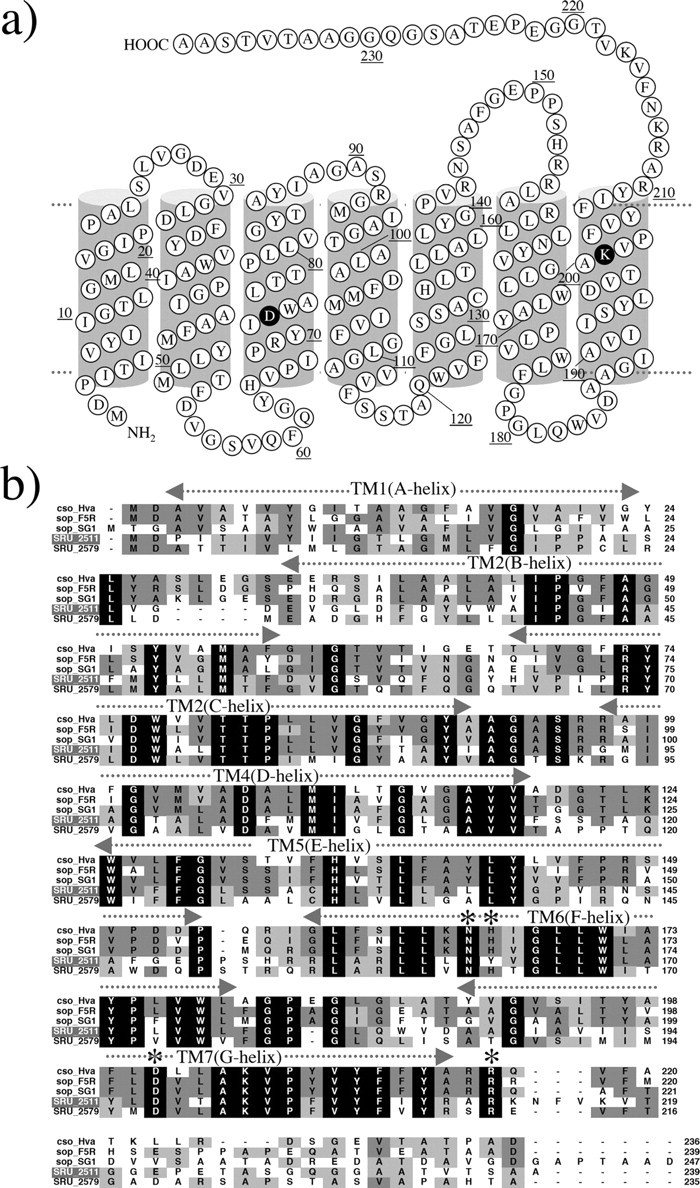
a, secondary structure of S. ruber sensory rhodopsin I (SrSRI, SRU_2511). b, alignment of putative amino acid sequences of SRIs reported so far (29) with SrSRI. Names of archaeal and bacterial sensory rhodopsins and sources from top are: cso_Hva, Haloarcula vallismortis cruxsensory rhodopsin (GenBank™ code: D83748); sop-F5R, H. salinarum strain Flx5R sR (L05603); sop-SGl, Hb. sp. strain SGI sR (X70290); SRU_2511, S. ruber strain DSM 13855 (3852586); SRU_2579, S. ruber strain DSM 13855 (3852268). Amino acid residues marked by an asterisk in helices F and G are important residues for phototaxis in SRI. The amino acid residues identified as necessary for function (31) were conserved among them except for a His residue (His-166 in HsSRI).
