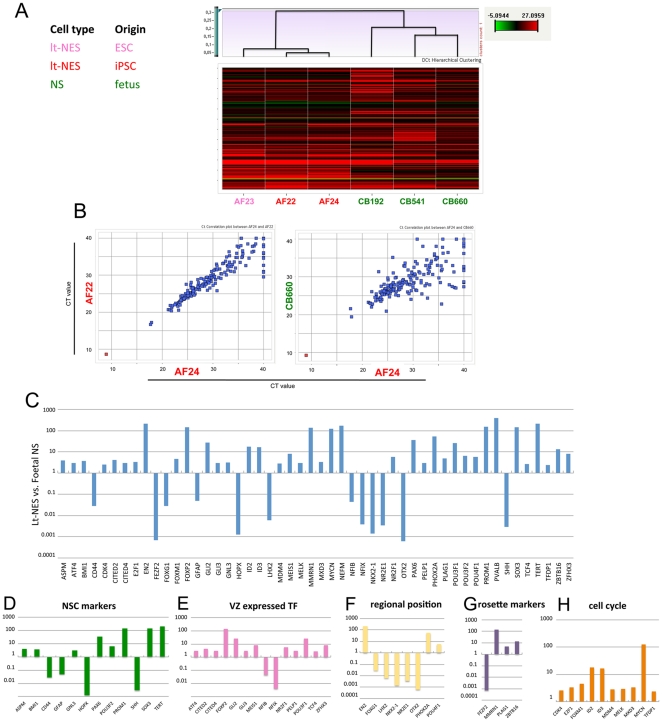
Figure 6. Gene expression analysis of lt-NES cells and fetal NS cells.
(A) A total of six cell lines representing two cell types (lt-NES and fetal NS) of different origins were profiled using TLDAs. Hierarchical cluster visualization (Pearson) shows separation into two major clusters representing the two cell types analyzed. (B) Correlation plots based on CT gene expression values of the individual cell lines. The lt-NES cell lines AF24 and AF22 show a higher degree of correlation than the lt-NES cell line AF24 and the NS line CB660, although the latter carry the same genome (R = 0.95 vs. R = 0.76). Red dots represent the median aggregated CT expression value of the 18S endogenous control; blue dots indicate the median aggregated CT expression values of all other genes on the TLDA. (C) Relative levels of differently expressed genes (corrected p-value threshold 0.05) in 9 lt-NES cell lines and 3 fetal NS cell lines. (D–H) Panels depicting the expression of markers representing the categories neural stem cell (NSC) markers, ventricular zone (VZ) expressed transcription factors (TF), regional position, rosette genes, and cell cycle. Data in C–H are depicted at a Log10 scale with the mean expression in fetal NS cells set to 1. No significant differences in expression levels were detected in ESC- vs. iPSC-derived lt-NES cells.