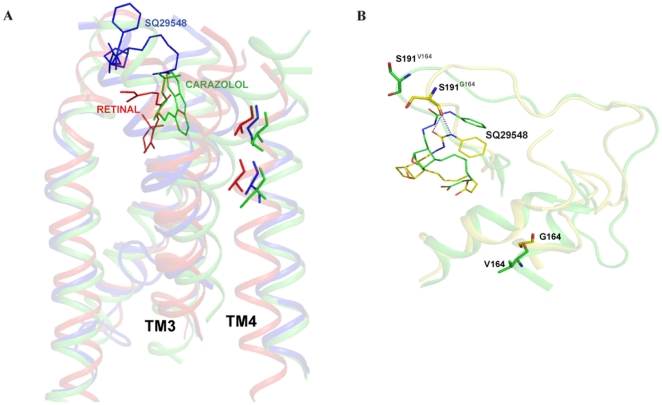
Figure 7. Molecular models of antagonist bound TP and G164V mutant and comparison with the crystal structures of rhodopsin and antagonist bound β2-AR.
Panel A, Molecular model of TP bound SQ 29,548 superimposed with the structures of rhodopsin (PDB ID 1U19) and antagonist bound β2-AR (PDB ID 2RH1). The two residues at positions 4.53 and 4.57 on TM4 in both rhodopsin and β2-AR were previously studied. For structural comparison all the three structures were superimposed. The color coding is as follows; Ala164 and Ala168 in rhodopsin (red), Ala160 and Gly164 in TP (blue) and Ser161 and Ser164 in β2-AR (green). Panel B, Molecular models of TP (yellow) and G164V (green) superimposed. The important amino acids Gly164, Val164 and Ser191 are shown. Notice that Ser191 loses interaction with the antagonist SQ 29,548 in the G164V mutant.