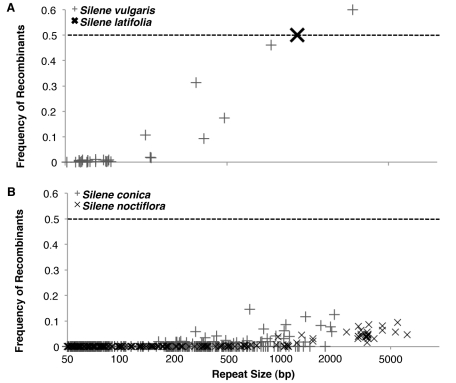
Figure 6. Repeat-mediated recombinational activity in the low mutation rate S. latifolia and S. vulgaris mitochondrial genomes (A) and the fast-evolving S. noctiflora and S. conica mitochondrial genomes (B).
Each point represents a pair of repeats, and its position on the y-axis denotes the proportion of recombinant genome conformations detected with paired-end 454 reads. The dashed lines indicate the level at which equal frequencies of read pairs support recombinant and nonrecombinant conformations. The S. latifolia mitochondrial genome was not sequenced with 454 paired-end reads, but Southern blot hybridizations indicated that alternative genome conformations associated with its six-copy 1.4-kb repeat exist at roughly equivalent frequencies [38], as indicated by the large X.