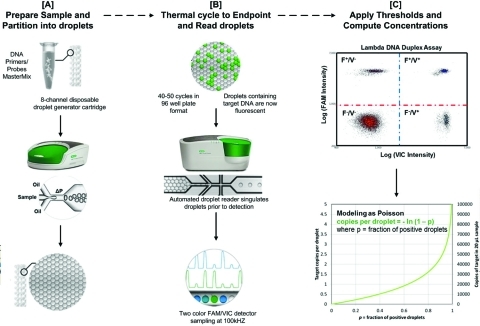
Figure 1.
Schematic showing the ddPCR workflow. (A) Each 20 μL sample containing the Master Mix, primers, TaqMan probes, and DNA target is loaded in the middle wells of a disposable eight channel droplet generator cartridge (pictured). Droplet generation oil (8 × 60 μL) containing the emulsion stabilizing surfactant is then loaded into the left-hand wells of the droplet generator cartridge. A vacuum is automatically applied to the outlet well (right) creating a pressure difference that, together with the geometry of the microfluidic circuit, converts the aqueous sample into stable, monodisperse, water-in-oil droplet emulsions which concentrate due to density differences from the oil phase and accumulate in the droplet collection wells of the cartridge. The droplets from each well are then transferred to one well of a 96-well plate, foil sealed, and thermal-cycled to the end-point. (B) After amplification, the plate is then loaded to a droplet reader where an autosampler aspirates the droplets and, using a microfluidic singulator, streams them single file (∼1500 droplets/s) past a FAM/VIC two color fluorescence detector which samples at a rate of 100 kHz. (C) The difference in fluorescence amplitudes for droplets where amplification has or has not occurred (positive and negatives, respectively) divides the entire droplet population into four discrete clusters for a typical Fam/Vic duplex assay. These four populations are droplets containing either no target (F–/V−), one of the targets (F–/V+, F+,V−), or both targets (F+,V+). Setting a fluorescence threshold for each detection channel affords a digital method of droplet classification and computing the average number of copies per droplet based on the fraction of positive droplets and Poisson modeling.