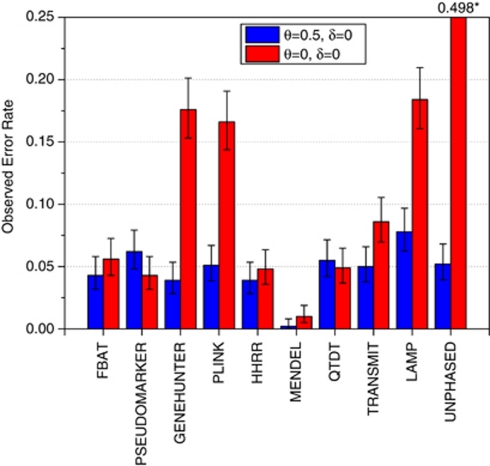
Figure 1.
Type-I error rates are presented for the author-recommended analysis options for testing for LD in the presence of linkage. Full details for all program options are in Supplementary Tables 2–5. The following analysis options were used from each program in this figure: FBAT's robust variance estimator and recessive model, PSEUDOMARKER MRec LD-given linkage, GENEHUNTER TDT, PLINK's sib-TDT, HHRR (allele-based randomized), MENDEL association given linkage using MRec*,fixed1, QTDT with no additional options, TRANSMIT's robust estimator and multiple nuclear families, LAMP's recessive model, and UNPHASED's ‘missing' and ‘parentrisk' options (with controls, cc). Blue columns represent empirical error rates estimated from the Finnish schizophrenia dataset – simulation of no linkage and no association. Other statistics from each program behaved similarly, with most being valid tests in this situation, with the notable exception of LAMP, and for LAMP the invalidity extended over the range of analysis models available in the program. The red columns are empirical error rates estimated from the Finnish schizophrenia dataset – simulation of complete linkage and no association. GENEHUNTER, PLINK, UNPHASED, and LAMP had elevated type-I error rates. Similar results were obtained for migraine families as well, and the pattern was the same for α=0.01. Results are based on 1000 replicates.