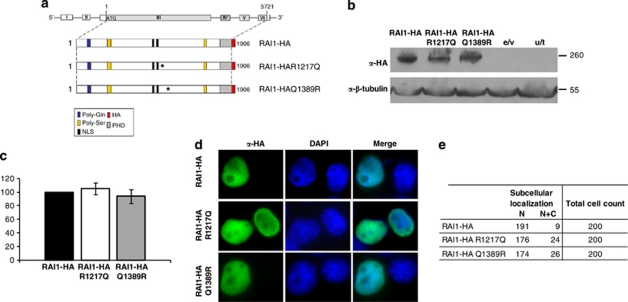
Figure 3.
In vitro evaluation of two point mutations associated with SMS. (a) Schematic representation of RAI1-HA, RAI1-HA R1217Q and RAI1-HA Q1389R. In blue is represented the poly-Gln domain, in yellow: poly-Ser domains, the in silico described nuclear localization signals are depicted in black, in slanted line: the PHD domain. The coding sequence for HA epitope is represented in red. Both generated mutants were confirmed by DNA sequencing (data not shown). (b) The molecular weight of mutated proteins was calculated by western blot analysis with anti-HA antibody in Neuro-2a cells extracts. The obtained molecular weight is depicted and also the controls for the immunoreactivity are shown (e/v, extracts transfected only with the empty vector and u/t represents untransfected cells control). Anti-β-tubulin was used as a loading control. (c) The percentage of the reporter transcription activation is shown for RAI1-HA R1217Q (white) and RAI1-HA Q1389R (gray) compared with RAI1-HA wild-type protein (black). Values represent mean±SEM. (d) Each plasmid was transfected in Neuro-2a cells and immunofluorescence was performed with anti-HA (green) antibody, whereas nuclei staining is shown with DAPI. (e) The subcellular localization of 200 positive cells for the immunodetection is summarized. α, Antibody against.