Abstract
Cementum has been shown to contain unique polypeptides that participate in cell recruitment and differentiation during cementum formation. We report the isolation of a cDNA variant for protein-tyrosine phosphatase-like (proline instead of catalytic arginine) member-a (PTPLA) from cementum. A cementifying fibroma-derived λ-ZAP expression library was screened by panning with a monoclonal antibody to cementum attachment protein (CAP), and 1435 bp cDNA (gb AC093525.3) was isolated. This cDNA encodes a 140-amino-acid polypeptide, and its N-terminal 125 amino acids are identical to those of PTPLA. This isoform, designated as PTPLA-CAP, results from a read-through of the PTPLA exon 2 splice donor site, truncating after the second putative transmembrane domain. It contains 15 amino acids encoded within the intron between PTPLA exons 2 and 3, which replace the active site for PTPLA phosphatase activity. The recombinant protein, rhPTPLA-CAP, has Mr 19 kDa and cross-reacts with anti-CAP antibody. Anti-rhPTPLA-CAP antibody immunostained cementum cells, cementum, heart, and liver. Quantitative RT-PCR showed that PTPLA was expressed in all periodontal cells; however, PTPLA-CAP expression was limited to cementum cells. The rhPTPLA-CAP promoted gingival fibroblast attachment. We conclude that PTPLA-CAP is a splice variant of PTPLA, and that, in the periodontium, cementum and cementum cells express this variant.
Keywords: cementum attachment protein, cementum, protein tyrosine phosphatase-like (proline instead of catalytic arginine), PTPLA, 3-hydroxyacyl-CoA dehydratase 1, periodontal ligament
Introduction
The cementum has been shown to contain polypeptides which have limited expression in cementum, cementoblasts, and putative cementoblasts. These polypeptides, which include cementum attachment protein (CAP) and cementum Protein 1 (CEMP1), are believed to regulate the biological activities of periodontal cells (Grzesik and Narayanan, 2002; Alvarez-Pérez et al., 2006). Characterization of the gene expression and structure of these molecules is essential to an understanding of the molecular mechanisms that control cementum formation and identification of specific markers for cementum (Bartold et al., 2000; Grzesik and Narayanan, 2002).
The CAP has been shown to play a role in cell recruitment and differentiation during cementum formation (Arzate et al., 1992a; Pitaru et al., 1995; Wu et al., 1996; Saito et al., 2001; Grzesik and Narayanan, 2002). It is expressed by dental follicle cells and promotes their adhesion and differentiation (Saito et al., 2001; Handa et al., 2002a,b; Kémoun et al., 2007), and periodontal cells that strongly bind CAP are able to form cementum-like mineralized tissue in culture (Liu et al., 1997; BarKana et al., 2000). Here we report the isolation of a cDNA using a monoclonal antibody raised against the CAP. This cDNA appears to be an alternative splice variant of protein tyrosine phosphatase-like (proline instead of catalytic arginine) member-a (PTPLA; Uwanogho et al., 1999; Li et al., 2000). The protein, designated as PTPLA-CAP, was expressed in bacteria. We show that the recombinant protein has properties similar to those of the CAP isolated from cementum tissue, that its expression is limited to a few tissues, and that, in the periodontium, it is expressed by cementoblastic cells.
Materials & Methods
Isolation of CAP cDNA and Expression of Recombinant Protein
A λ-ZAP Express EcoRI/XhoI (Stratagene, La Jolla, CA, USA) cDNA library was constructed with mRNA from human cementum tumor-derived cells (Arzate et al., 1992b). After transient expression in cos-7 cells, it was screened by panning with monoclonal anti-CAP antibody 3G9 (Saito et al., 2001). Panning plates were prepared as described elsewhere (Alvarez-Pérez et al., 2006), and positives were analyzed by PCR with T3 and T7 primers. Recombinant clones were constructed with a pENTR/SD/D-TOPO vector for directional cloning of a blunt-end PCR product (Invitrogen, Carlsbad, CA, USA). For protein expression, PCR product was obtained with sense primer 5′ CACC ATG GGG CGC CTG ACG GAA GCG 3′, and antisense primer 5′ CAT AAA TAT TAC AGC AAT AGA 3′; we designed this sequence by eliminating the native stop codon to fuse the PCR product in frame with the COOH terminal tag of the entry clone in a Gateway pET-DEST 42 vector (Invitrogen) for inducible expression. Protein was expressed in Escherichia coli BL21 strain (Invitrogen) in medium containing 100 μg/mL ampicillin (Alvarez-Pérez et al., 2006). Recombinant protein (rhPTPLA-CAP) was purified from bacterial cell lysates by Ni2+ affinity chromatography HisPrep FF 16/10 column (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA), followed by hydroxyapatite (HA)-Ultrogel affinity chromatography (Sigma, St. Louis, MO, USA).
Antibody Preparation
Hydroxyapatite affinity-purified recombinant rhPTPLA-CAP was separated on 12% SDS-PAGE gel, and Mr 19,000 band was used to immunize New Zealand rabbits (Alvarez et al., 2003).
Cells and Determination of Attachment Activity
Human gingival fibroblasts, cementoblasts, periodontal ligament cells, and alveolar bone cells were obtained as described previously (Arzate et al., 1992b; Pitaru et al., 1995). Attachment activity was determined with human gingival fibroblasts in plates coated with rhPTPLA-CAP (Pitaru et al., 1995; Wu et al., 1996). Wells coated with fibronectin served as positive control, and serum-free medium was negative control.
SDS-Polyacrylamide Gel Electrophoresis and Western Blot
Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE, 12% gels; for protein extraction from tissues and other details, please see the Appendix). Western blot analyses were performed with monoclonal anti-His (C-term) antibody (Invitrogen), monoclonal anti-bovine CAP antibody 3G9 (Saito et al., 2001; Santa Cruz Biotech., Santa Cruz, CA, USA), and polyclonal anti-rhPTPLA-CAP antibody, as described previously (Wu et al., 1996; Saito and Narayanan, 1999).
Immunostaining
Human tissues used were obtained from the Department of Pathology, School of Dentistry, UNAM, in conformity with the policy of the Research Review Board, Facultad de Odontología, UNAM. Formalin-fixed 5-μ sections were incubated with antigen retrieval solution (Shi et al., 1992) and immunostained (Arzate et al., 1998). Cells were plated at a density of 0.5 x 103cells/well in 8-well Lab-Tek chamber slides, cultured for 3 days, and immunostained as described previously (Arzate et al., 1992a; Alvarez-Pérez et al., 2006). Negative controls were sections immunostained with pre-immune rabbit serum or without primary antibody.
Quantitative Real-time PCR (qRT-PCR)
Total RNA was isolated with the Trizol Reagent (Invitrogen) and amplified by PCR with SuperScript™ III Platinum® SYBR® Green One-Step qRT-PCR Kit with ROX (Invitrogen). Sense and antisense primers for PTPLA gene, respectively, were: 5′ ACTG GCT CAC CTT CTA CGA 3′ and 5′ GCA AGG CAA ATG TCT GGA 3′. PTPLA-CAP specific primers were: 5′ TCC AGA CAT TTG CCT TGC TT 3′ (sense); and 5′ TTA CAG CAA TAG AAA AAC AGC ATG A 3′ (antisense). Gyceraldehyde-3-phosphate (GAPDH) gene expression was used for normalization. A 25-μL reaction was set up with the following PCR conditions: (cDNA synthesis) 50°C for 3 min, denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 30 sec, and finally 40°C, 1 min. Amplifications were performed in a Corbett Rotor-Gene 6000 (Qiagen, Valencia, CA, USA). All experiments were performed in triplicate and normalized for GAPDH expression.
Statistical Analysis
Assays were performed in triplicate and repeated twice. Statistical significance was determined by Student’s t test with Sigma Stat V 3.1 software. Significance level was set at alpha = 0.05.
Results
Sequence Analysis
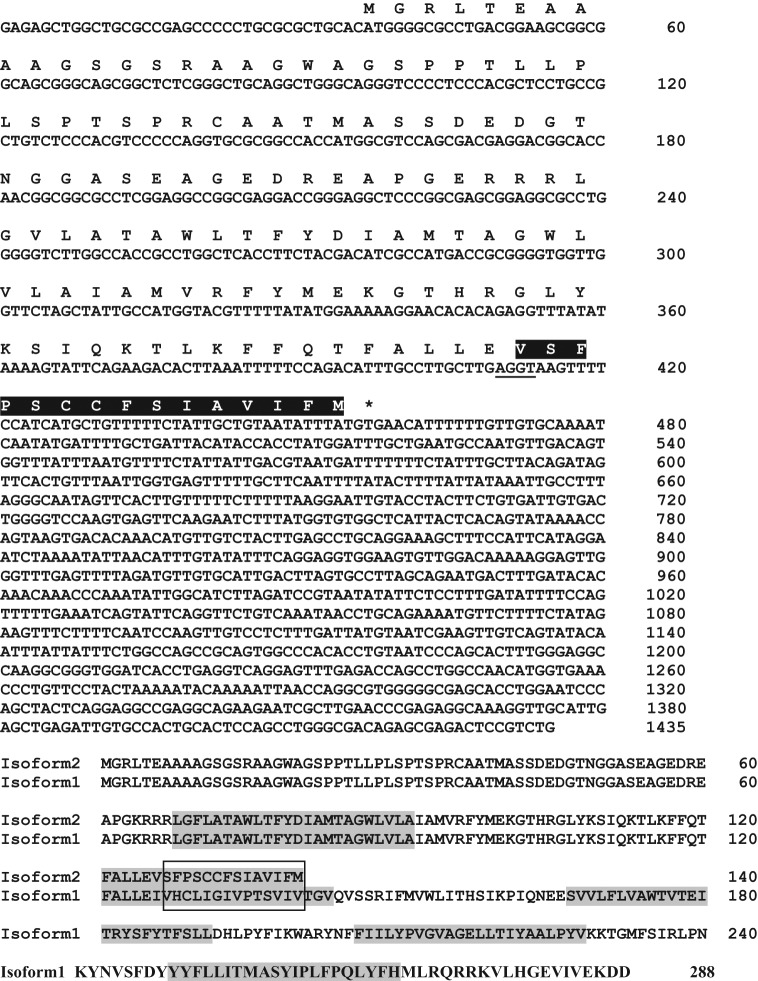
The human cementifying fibroma λ-Zap expression cDNA library was transiently expressed in cos-7 cells. After 3 rounds of “panning” with anti-bovine CAP IgG antibody (3G9), a 1435-bp cDNA was isolated. The nucleotide and deduced amino acid sequences of this cDNA are shown in Fig. 1. It has a consensus Kozak initiation sequence (CXXATGG), and the translation start site (ATG) is at nucleotide 37. The stop codon TGA is at base 457, and this is followed by a 3′ untranslated region of 979 nucleotides. The cDNA, designated tentatively as PTPLA-CAP, encodes a polypeptide of 140 amino acids (Fig. 1; GenEMBL AY455942; gb AC093525.3). BLAST analysis of the non-redundant NCBI protein database revealed that the N-terminal 125 amino acids of PTPLA-CAP and 3-hydroxyacyl-CoA dehydratase 1/protein tyrosine phosphatase-like (proline instead of catalytic arginine) gene encoded on chromosome 10p13–p14 (NP_055056) are identical. A T-BLASTN search of the human genome demonstrated that the C-terminal 15 amino acids of the PTPLA-CAP are not present in PTPLA sequence (isoform 1, Fig. 1), and it is encoded by a read-through of the splice donor site of exon 2 that is utilized for expression of the full-length PTPLA with 288 amino acids (Uwanogho et al., 1999; Li et al., 2000). Thus, the PTPLA-CAP represents a second isoform of the 3-hydroxyacyl-CoA dehydratase 1/PTPLA. The PTPLA (isoform 1; Fig. 1) is predicted to contain 4 to 5 transmembrane spanning domains, and the second putative transmembrane domain is encoded by exon 2 (residues 121-125) and exon 3 (residues 126-143). Residues 126-136 of the second transmembrane domain contain the PTPLA active site IVHCLIGIVPT and the catalytic cysteinyl residue. In PTPLA-CAP, the exon 3 residues are replaced by the 15-amino-acid sequence VSFPSCCFSIAVIFM encoded by read-through into the intron downstream of exon 2 (Fig. 1). These intron-encoded 15 residues are hydrophobic and provide the remainder of the residues necessary for the second transmembrane domain initiated within exon 2. The PTPLA-CAP protein truncates immediately after the second transmembrane domain and lacks the active site for phosphatase activity of PTPLA. Sequence analysis (SignalP 3.0 server) shows only a low probability of the presence of signal sequence, with probability of cleavage between positions 38 and 39.
Figure 1.
The cDNA and deduced amino acid sequence of PTPLA-CAP (AY455942). In PTPLA-CAP, the sequence VSFPSCCFSIAVIFM, which replaces IVHCLIGIVPT in PTPLA (isoform 1; AF114494), is highlighted in black. For a schematic drawing showing mRNA sequence and exon positions of PTPLA-CAP and PTPLA, see Schild et al. (2009). Isoform 2 encoding the PTPLA-CAP protein is encoded by an alternate spliced mRNA that does not have a splice between exons 2 and 3; instead, the C-terminal end of the protein terminates after 14 residues encoded within the intron between exons 2 and 3. The non-functional splice donor site “AGGT” of exon 2 is underlined. The predicted transmembrane spanning domains in PTPLA (see isoform 1) are highlighted in gray. The 14 intron encoded amino acids (boxed) constitute the C-terminal half of a second putative transmembrane domain within the PTPLA-CAP protein; thus, the alternately spliced isoform 2 (PTPLA-CAP protein) lacks the additional transmembrane domains in the longer isoform 1 PTPLA. Sequence analysis shows only a low probability of a signal sequence, with cleavage probability between positions 38 and 39.
The predicted translation product has a calculated molecular weight 14.9 kDa and pI 7.72. Analysis of predicted post-translational modifications (ExPASy) revealed that it does not contain N-glycosylation sites, while potential O-glycosylation motifs (S/T) are located at positions 5, 22, 25, 30, 32, 33, 39, 42, 43, and 48. Theoretical potential phosphorylation motifs, including a cdc2 (position 12, 22, 32), cdk5 (position 22, 30, 32), GSK3 (position 30), p38 MAPK (position 30, 33), CKII (position 42, 43, 53), and PKC (position 114), are present.
Purification of Recombinant Human PTPLA-CAP
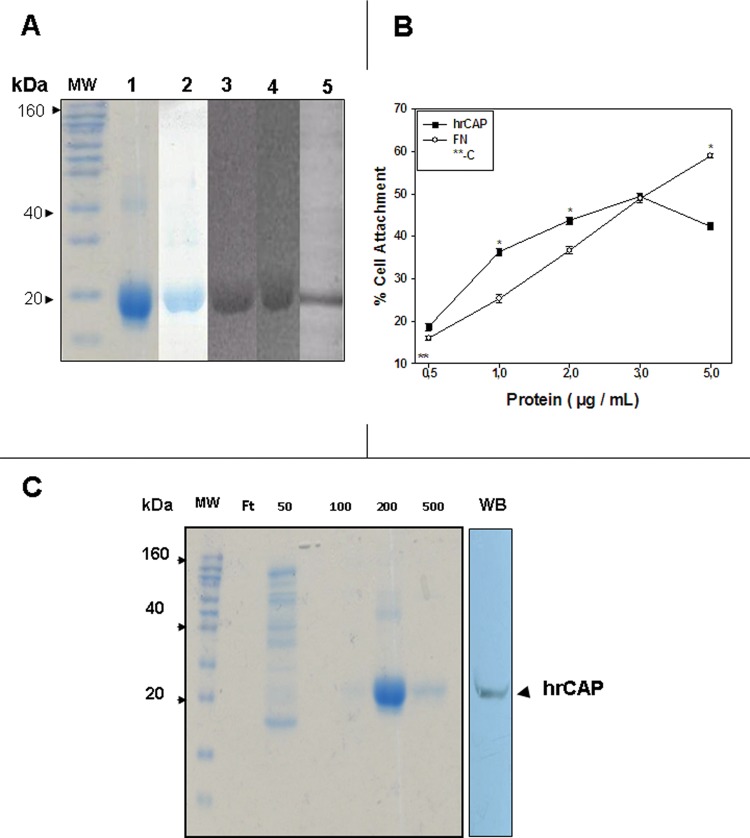
The protein was expressed in E. coli and purified with Ni2+ and hydroxyapatite affinity chromatography. The purified protein migrated with ~19 kDa in SDS-polyacrylamide gels (lanes 1 and 2, Fig. 2A). It was recognized by monoclonal anti-His (C-term) (lane 3) and polyclonal anti-rhPTPLA-CAP (lane 5) antibodies, and by anti-CAP monoclonal antibody 3G9 (lane 4, Fig. 2A).
Figure 2.
Properties of rhPTPLA-CAP. SDS-polyacrylamide gel electrophoresis of proteins expressed by bacteria carrying the expression construct. Lane 1: Novex Sharp (Invitrogen) protein markers, Coomassie-blue-stained. Lane 2: expressed protein, before purification, Coomassie-blue-stained. Lane 3: expressed protein, after Ni2+ affinity chromatography. Lane 4: Western blot, anti-6xHis antibody. Lane 5: Western blot, anti-CAP antibody 3G9. Lane 6: Western blot, anti-rhPTPLA-CAP polyclonal antibody. (B) Attachment assay with purified rhPTPLA-CAP. Human gingival fibroblasts were plated at 2 x 104 cells/well in 24-multiwell Costar plates not treated for tissue culture and previously coated with 0.5, 1.0, 2.0, and 5.0 mg/mL of hrPTPLA-CAP. Cell attachment was measured after 4 hrs. Wells coated with 5 mg/mL of fibronectin served as positive control (FN), and serum-free medium was negative control (**C). Values plotted represent mean ± SD of triplicates. * indicates statistically significant differences (p < 0.05). (C) Binding of rhPTPLA-CAP to hydroxyapatite (HA). The recombinant protein was loaded on a HA Ultrogel column and eluted with 50, 100, 200, and 500 mM Na2PO4, pH 7.2. Coomassie blue staining shows that the rhPTPLA-CAP is eluted by 200-500 mM Na2PO4. Ft: flow through. Western blot (WB) shows that the HA-binding protein (hrCAP) cross-reacts with anti-bovine CAP monoclonal antibody (3G9; arrow).
Cell Attachment Activity
The rhPTPLA-CAP promoted attachment of human gingival fibroblasts, and the attachment was dependent upon protein concentration (Fig. 2B). The attachment activity was comparable with that of fibronectin.
Immunostaining and Localization of CAP
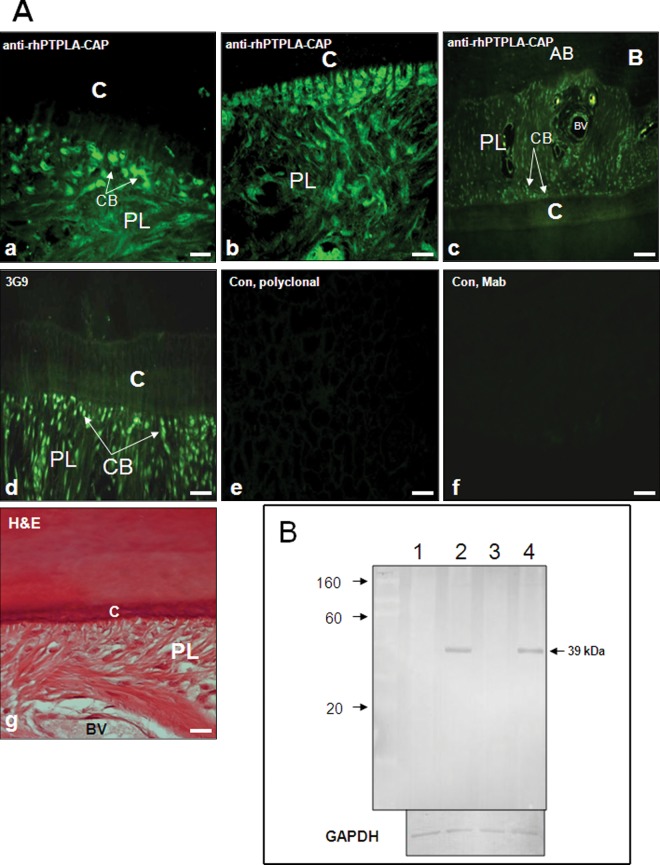
Immunostaining with anti-rhPTPLA-CAP antibody showed staining of cementoblasts facing cementum and the matrix (Fig. 3a). Collagen-like fibers of periodontal ligament and periodontal ligament cell population were stained positively (Figs. 3b, 3c). The staining was similar to that of anti-CAP monoclonal antibody 3G9, which cross-reacts with cementoblasts and the periodontal ligament cell population (Fig. 3d). Cementoblastoma-derived cells were strongly positive, while periodontal ligament cells were moderately positive, and alveolar-bone-derived cells and human gingival fibroblasts were negative (Appendix Fig. 1A). Heart, liver, and kidney showed positive staining, and other tissues had weak or no reactivity (Appendix Fig. 1B). Western blot analysis detected a ~39 kDa protein in extracts of cementum and periodontal ligament tissues, but not gingiva and alveolar bone (Fig. 3B).
Figure 3.
Expression of PTPLA-CAP in human periodontium.(A) Immunostaining of periodontal tissue with anti-rhPTPLA-CAP polyclonal antibody. Staining is observed in cementoblasts facing cementum and in the matrix (a) (arrows). Cementoblasts and collagen-like fibers of the periodontal ligament are stained positively (b) Expression of PTPLA-CAP in human periodontium. Lower view of periodontal tissue shows immunostaining in cementoblasts (arrows) and periodontal ligament cell population (c). Monoclonal antibody against bovine cementum attachment protein (3G9) cross-reacts with cementoblasts (arrows) and periodontal ligament cell populations (d). Controls with pre-immune rabbit (e) or mouse serum (f) were negative. H&E staining shows tissue orientation (g). BV, blood vessel; C, cementum; PL, periodontal ligament. Magnification 20x (Bar – 100 mm). (B) Western analysis of gingival tissue (lane 1), cementum (lane 2), alveolar bone (lane 3), and periodontal ligament (lane 4) extracts with anti-rhPTPLA-CAP antibody. The antibody recognizes a 39-kDa protein in extracts of gingival and periodontal ligament tissues. The bottom panel shows staining for GAPDH for protein loading.
Expression in Periodontal Cells
Immunostaining showed the presence of PTPLA-CAP in cementum and cementum cells (Fig. 3); in contrast, PTPLA is expressed in many tissues (Uwanogho et al., 1999; Li et al., 2000), but not in rat teeth (Schild et al., 2009). Therefore, we compared the expression of PTPLA-CAP and PTPLA in human periodontal cells. We performed Quantitative PCR using primer pairs for PTPLA and for the sequence flanking the alternatively spliced domain in PTPLA-CAP (Fig. 1). The results showed that PTPLA was expressed in gingival fibroblasts, periodontal ligament cells, and osteoblasts, and it was expressed relatively less (~10% relative to gingival fibroblasts) in cementum cells (Fig. 4A). In contrast, the alternatively spliced PTPLA-CAP sequence was expressed only in cementoblasts (Fig. 4B).
Figure 4.
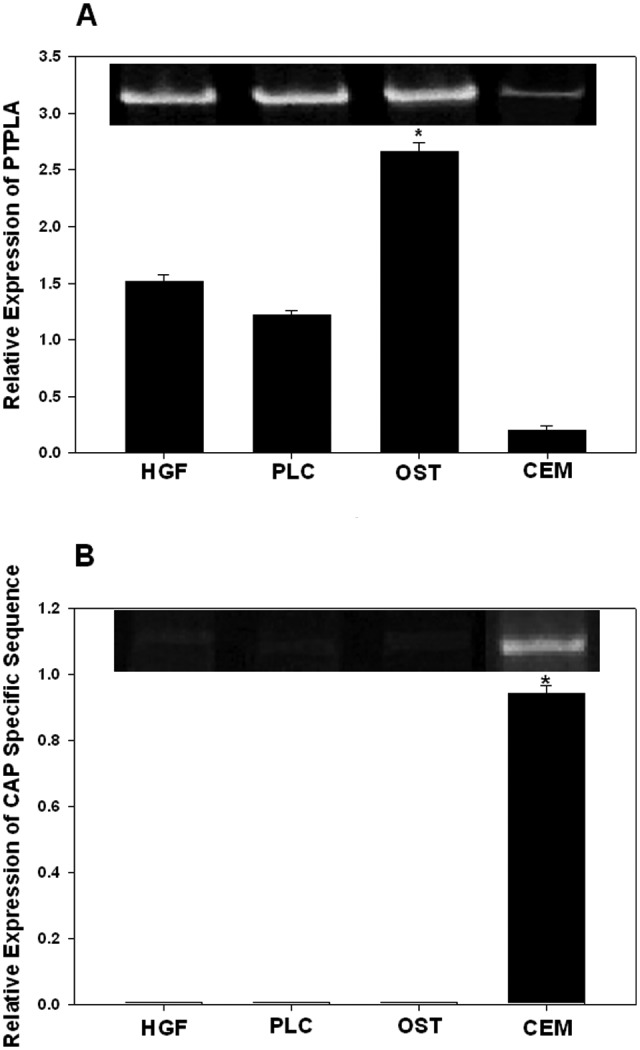
Relative expression of PTPLA and PTPLA-CAP determined by quantitative PCR after normalizing for GAPDH. (A) PTPLA is expressed in human gingival fibroblasts (HGF), periodontal ligament cells (PLC), and alveolar-bone-derived cells (OST). Cementoblastoma-derived cells showed relatively weak expression. No bands were visible in controls without reverse transcriptase (data not shown). (B) Expression of PTPLA-CAP measured with the primer sequences specific for PTPLA-CAP. Values plotted in A and B represent mean ± SD of triplicates. *Differences were statistically significant (p < 0.005).
Discussion
Our study has identified an alternately spliced mRNA that encodes a truncated isoform of 3-hydroxyacyl-CoA dehydratase 1/PTPLA. This mRNA, PTPLA-CAP, encodes a 140-amino-acid protein that is identical to the first 125 N-terminal amino acids of PTPLA, which is 288 amino acids long (isoform 1, Fig. 1). The remainder of the C-terminus of PTPLA-CAP is encoded by a read-through of the splice donor site in exon 2 of the PTPLA isoform into the adjacent intron. This protein is predicted to have 2 transmembrane spanning domains, and it truncates immediately after the second transmembrane domain. The truncation eliminates the PTPLA sequence IVHCLIGIVPT, which has the signature phosphatase active site motif (I/VHCX XGXXP(S/T) (Uwanogho et al., 1999; Li et al., 2000), and additional C-terminal transmembrane spanning domains predicted for the PTPLA isoform. We have examined the NCBI EST database for the presence of corresponding EST sequences and detected 1 human and 1 rat EST showing the same alternate splice. The human EST (CN305861) was derived from undifferentiated human embryonic stem cell lines maintained on a feeder-free layer.
The PTPLA mRNA is widely expressed in many tissues (Uwanogho et al., 1999; Li et al., 2000; Schild et al., 2009). In the periodontium, PTPLA is expressed in gingival fibroblasts, periodontal ligament cells, and osteoblasts, and only marginally in cementum cells (Fig. 4); in contrast, qRT-PCR revealed that the PTPLA-CAP is expressed in cementum cells, not in other cells (Figs.3A, Figs.4). It is also expressed in the heart. This result is consistent with a recent report demonstrating expression in rat heart and muscle (Schild et al., 2009). This report demonstrates no expression in rat teeth, and our results showed a low level of expression in cementum cells (Fig. 4); the reasons for this difference are not clear, although one possibility is the human vs. rat species difference. It is likely that the low level of PTPLA expressed in cementum cells (Fig. 4A) represents the PTPLA-CAP in these cells.
The rhPTPLA-CAP migrates with ~19 kDa, while the anti-rhPTPLA-CAP antibody recognizes a 39-kDa protein in cementum and periodontal ligament extracts. We believe that the most likely explanation for the size difference between the recombinant and cell/tissue proteins is post-translational modification; we have not performed experiments to determine this possibility. Cell lysates of gingival fibroblasts, periodontal ligament cells, bone cells, and cementum cells contain ~40 and 38 kDa species; these are absent in blots with antibody pre-incubated with rhPTPLA-CAP (Appendix Fig. 2), indicating that these represent PTPLA-CAP. The PTPLA-CAP is not detected in gingiva and bone (Fig. 3A), while it is detectable in cells of these tissues; one possible reason for this difference is that in vitro culture conditions facilitate the expression and persistence of PTPLA-CAP in culture.
The CAP promotes the preferential adhesion of putative cementoblasts (Liu et al., 1997; BarKana et al., 2000; Saito et al., 2001). Adhesion to CAP is sufficient to promote growth-factor-induced DNA synthesis, and this is mediated through signaling events participating in cell- cycle events (Saito and Narayanan, 1999; Komaki et al., 2000; Yokokoji and Narayanan, 2001). The action of CAP appears to be mediated through binding to α5β1 integrin (Ivanovski et al., 1999); however, we do not know if the rhPTPLA-CAP also binds to this receptor. Like the CAP, the rhPTPLA-CAP promotes attachment of periodontal cells and binds to hydroxyapatite (Pitaru et al., 1992) (Fig. 2C). These observations indicate that the PTPLA-CAP and CAP may play a regulatory role during cementum formation (Grzesik and Narayanan, 2002). However, the significance of its homology to PTPLA and 3-hydroxyacyl-CoA dehydratase is intriguing. The enzyme 3-hydroxyacyl-CoA dehydratase (HACD1/PTPLA) and 3 other homologues are involved in the third step, catalyzing the dehydration of 3-hydroxyacyl-CoA during the synthesis of very long-chain fatty acids (Ikeda et al., 2008); whether the CAP has a similar function is not clear. The homologue of PTPLA-CAP, PTPLA, is a phosphatase; however, its active site is absent in PTPLA-CAP. Thus, one possibility is that the PTPLA-CAP may regulate the phosphatase activity of PTPLA. Interestingly, cyclosporine A, which inhibits calcineurin, another phosphatase, also induces cementum formation and mineralization (Ayanouglou, 1998; Arzate et al., 2005). If this is the case, the outcome of PTPLA-CAP action is likely to be different in other tissues, which are not mineralized, where it is expressed. Further work is necessary to investigate these possibilities.
Acknowledgments
We are indebted to Dr. Tim Rose, Department of Pediatrics, University of Washington and Seattle Children’s Research Institute, for his help with sequence analysis.
Footnotes
This work was supported by NIH grants DE 13069 and DE-08229, DGAPA-UNAM-PAPIIT IN216711, and CONACyT 152206 and 130950. We thank Melinda Ng and Hanah Kim for technical assistance.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Alvarez MA, Pitaru S, Alvarez-Fregoso O, Reyes-Gasga J, Arzate H. (2003). Anti-cementoblastoma-derived protein antibody partially inhibits mineralization on a cementoblastic cell line. J Struct Biol 143:1-13 [DOI] [PubMed] [Google Scholar]
- Alvarez-Pérez MA, Narayanan S, Zeichner-David M, Rodríguez Carmona B, Arzate H. (2006). Molecular cloning, expression and immunolocalization of a novel human cementum-derived protein (CP-23). Bone 38:409-419 [DOI] [PubMed] [Google Scholar]
- Arzate H, Olson SW, Page RC, Gown AM, Narayanan AS. (1992a). Production of a monoclonal antibody to an attachment protein derived from human cementum. FASEB J 6:2990-2995 [DOI] [PubMed] [Google Scholar]
- Arzate H, Olson SW, Page RC, Narayanan AS. (1992b). Isolation of human tumor cells that produce cementum proteins in culture. Bone Miner 18:15-30 [DOI] [PubMed] [Google Scholar]
- Arzate H, Alvarez M, Aguilar-Mendoza ME, Alvarez O. (1998). Human cementum tumor cells have different features from human osteoblastic cells in vitro. J Periodontal Res 33:249-258 [DOI] [PubMed] [Google Scholar]
- Arzate H, Alvarez MA, Narayanan AS. (2005). Cyclosporin-A promotes mineralization by human cementoblastoma-derived cells in culture. J Periodontal Res 40:218-224 [DOI] [PubMed] [Google Scholar]
- Ayanouglou CM. (1998). Evidence that cyclosporin-A administration induces the formation of new cementum-like islets inside the gingival connective tissue. J Periodontal Res 33:166-171 [DOI] [PubMed] [Google Scholar]
- BarKana I, Narayanan AS, Grosskop A, Savion N, Pitaru S. (2000). Cementum attachment protein enriches putative cementoblastic populations on root surfaces in vitro. J Dent Res 79:1482-1488 [DOI] [PubMed] [Google Scholar]
- Bartold PM, McCulloch CA, Narayanan AS, Pitaru S. (2000). Tissue engineering. A new paradigm for periodontal regeneration based on molecular and cell biology. Periodontol 2000 24: 253-269 [DOI] [PubMed] [Google Scholar]
- Grzesik WJ, Narayanan AS. (2002). Cementum and periodontal wound healing and regeneration. Crit Rev Oral Biol Med 13:474-484 [DOI] [PubMed] [Google Scholar]
- Handa K, Saito M, Tsunoda A, Yamauchi M, Hattori S, Sato S, et al. (2002a). Progenitor cells from dental follicle are able to form cementum matrix in vivo. Connect Tissue Res 43:406-408 [DOI] [PubMed] [Google Scholar]
- Handa K, Saito M, Yamauchi M, Kiyono T, Sato S, Teranaka T, et al. (2002b). Cementum matrix formation in vivo by cultured dental follicle cells. Bone 31:606-611 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Kanao Y, Yamanaka M, Sakuraba H, Mizutani Y, Igarashi Y, et al. (2008). Characterization of four mammalian 3-hydroxyacyl-CoA dehydratases involved in very long-chain fatty acid synthesis. FEBS Lett 582:2435-2440 [DOI] [PubMed] [Google Scholar]
- Ivanovski S, Komaki M, Bartold PM, Narayanan AS. (1999). Periodontal-derived cells attach to cementum attachment protein via α5β1 integrin. J Periodontal Res 34:154-159 [DOI] [PubMed] [Google Scholar]
- Kémoun P, Laurencin-Dalicieux S, Rue J, Farges JC, Gennero I, Conte-Auriol F, et al. (2007). Human dental follicle cells acquire cementoblast features under stimulation by BMP-2/-7 and enamel matrix derivatives (EMD) in vitro. Cell Tissue Res 329:283-294 [DOI] [PubMed] [Google Scholar]
- Komaki M, Kang M, Narayanan AS. (2000). Role of MAP kinases p42erk-2/p44erk-1 in cementum-derived attachment protein mediated cell attachment. J Dent Res 79:1789-1793 [DOI] [PubMed] [Google Scholar]
- Li D, Gonzalez O, Bachinski LL, Roberts R. (2000). Human protein tyrosine phosphatase-like gene: expression profile, genomic structure, and mutation analysis in families with ARVD. Gene 256:237-243 [DOI] [PubMed] [Google Scholar]
- Liu HW, Yacobi R, Savion N, Narayanan AS, Pitaru S. (1997). Collagenous cementum-derived attachment protein is a marker for progenitors of the mineralized tissue-forming cell lineage of the periodontal ligament. J Bone Miner Res 12:1691-1699 [DOI] [PubMed] [Google Scholar]
- Pitaru S, Savion N, Hekmati H, Olsen S, Narayanan AS. (1992). Binding of a cementum attachment protein to extracellular matrix components and to dental surfaces. J Periodontal Res 27:640-646 [DOI] [PubMed] [Google Scholar]
- Pitaru S, Narayanan AS, Olsen S, Savion N, Hekmati H, Alta I, et al. (1995). Specific cementum attachment protein enhances selectively the attachment and migration of periodontal cells to root surfaces. J Periodontal Res 30:360-368 [DOI] [PubMed] [Google Scholar]
- Saito M, Narayanan AS. (1999). Signaling reactions induced in human fibroblasts during adhesion to cementum-derived attachment protein. J Bone Miner Res 14:65-72 [DOI] [PubMed] [Google Scholar]
- Saito M, Iwase M, Maslan S, Nozaki N, Yamauchi M, Handa K, et al. (2001). Expression of cementum-derived attachment protein in bovine tooth germ during cementogenesis. Bone 29:242-248 [DOI] [PubMed] [Google Scholar]
- Schild C, Beyeler M, Lang NP, Trueb B. (2009). Cementum attachment protein/protein-tyrosine phosphatase-like member A is not expressed in teeth. Int J Mol Med 23:293-296 [PubMed] [Google Scholar]
- Shi SR, Coté C, Kalra KL, Taylor CR, Tandon AK. (1992). A technique for retrieving antigens in formalin-fixed, routinely acid-decalcified, celloidin-embedded human temporal bone sections for immunohistochemistry. J Histochem Cytochem 40:787-792 [DOI] [PubMed] [Google Scholar]
- Uwanogho DA, Hardcastle Z, Balogh P, Mirza G, Thornburg KL, Ragoussis J, et al. (1999). Molecular cloning, chromosomal mapping, and developmental expression of a novel protein tyrosine phosphatase-like gene. Genomics 62:406-416 [DOI] [PubMed] [Google Scholar]
- Wu D, Ikezawa K, Parker T, Saito M, Narayanan AS. (1996). Characterization of a collagenous cementum-derived attachment protein. J Bone Miner Res 11:686-692 [DOI] [PubMed] [Google Scholar]
- Yokokoji T, Narayanan AS. (2001). Role of D1 and E cyclins in cell cycle progression of human fibroblasts adhering to cementum attachment protein. J Bone Miner Res 16:1062-1067 [DOI] [PubMed] [Google Scholar]