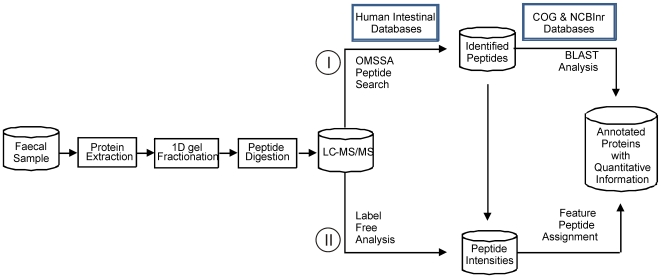
Figure 1. Metaproteomic analysis pipeline.
Proteins were released from cells by mechanical disruption and then separated on a 1D gel prior to LC-MS/MS analysis. The MS/MS spectra were searched with OMSSA against databases (see Table 3) containing sequences to be expected in faecal material. All MS spectra were aligned over different runs and peptide identification was assigned to the detected features (route I). To functionally annotate the identified peptides and proteins, BLAST analyses were performed against the COG and NCBI non-redundant (nr) databases (route II).