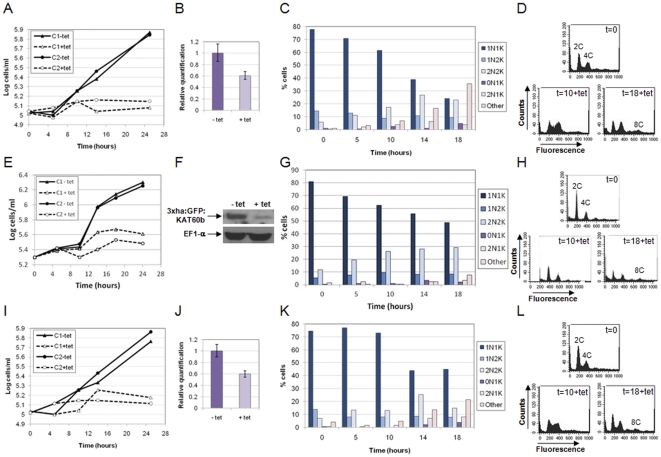
Figure 2. KAT60a, KAT60b and KAT60c are each essential for cytokinesis in bloodstream stage T. brucei.
Panels A–D: KAT60a; panels E–H: KAT60b; panels I–L: KAT60c. A, E, I: cumulative growth curves of two independent bloodstream RNAi clones (C1: clone 1; C2: clone 2) cultured in the presence or absence of tetracycline (tet). B,F,J: real time PCR (KAT60a and KAT60c) or Western analysis (KAT60b), at 8 hours post-induction showing downregulation of the relevant transcript or protein (data presented for clone 1 in each case). Error bars represent standard deviations. For KAT60b RNAi cell lines (F), one allele of KAT60b was replaced with 3xha:GFP:KAT60b to allow visualisation of KAT60b downregulation by Western blotting with anti-HA antibody. The membrane was also probed with anti-elongation factor 1 alpha (EF1-α) antisera as a loading control. C, G, K: DAPI staining. The nucleus (N) and kinetoplast (K) configurations of >200 cells per time point following RNAi induction of clone 1 are shown. ‘Other’ comprises mainly multinucleate and/or multi-kinetoplast cells. D, H, L: DNA content analysis. The fluorescence of 10,000 propidium iodide-stained cells was analysed by flow cytometry in the FL2-A channel at the time points (in hours) indicated for KAT60a, KAT60b and KAT60c RNAi cell lines. The ploidies of peaks are given. Data for clone 2 are comparable (not shown).