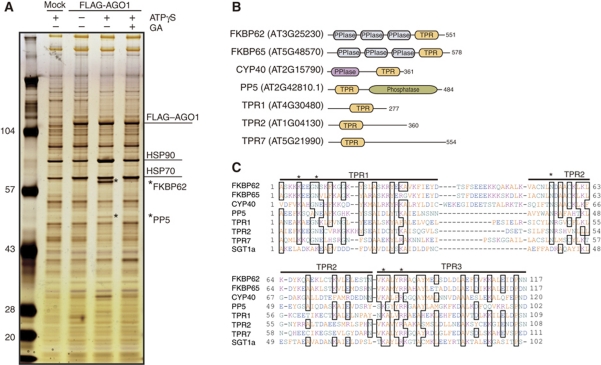
Figure 1.
Identification of factors that associate with AGO1. (A) Analysis of factors that are copurified with AGO1. FLAG–AGO1 mRNA-translated BYL was incubated at 25°C for 30 min in the presence of additional 2 mM ATP, 3 mM MgCl2, and 2% DMSO (−ATPγS –GA); 2 mM ATPγS, 3 mM MgCl2, and 2% DMSO (+ATPγS –GA); or 2 mM ATPγS, 3 mM MgCl2, 2% DMSO, and 20 μM GA (+ATPγS +GA). In parallel, mock-translated BYL was incubated in the +ATPγS –GA condition. FLAG–AGO1 was immunopurified with anti-FLAG antibodies, and copurified proteins were analysed by SDS–PAGE followed by silver staining. LC–MS/MS analysis identified two FLAG–AGO1-associated proteins corresponding to bands that were found only in the +ATPγS –GA condition (indicated with asterisks) to be FKBP62 and PP5. (B) Schematic representation of the domain structures of the TPR proteins. (C) Alignment of the amino-acid sequences of the TPR domains from A. thaliana proteins. The amino-acid sequences of the TPR domains of Arabidopsis FKBP62, FKBP65, CYP40, PP5, TPR1, TPR2, TPR7, and SGT1a were aligned using Clustal W (DNA Data Bank of Japan). Asterisks indicate the conserved residues required for interaction with the EEVD motif of HSP90 or HSP70.