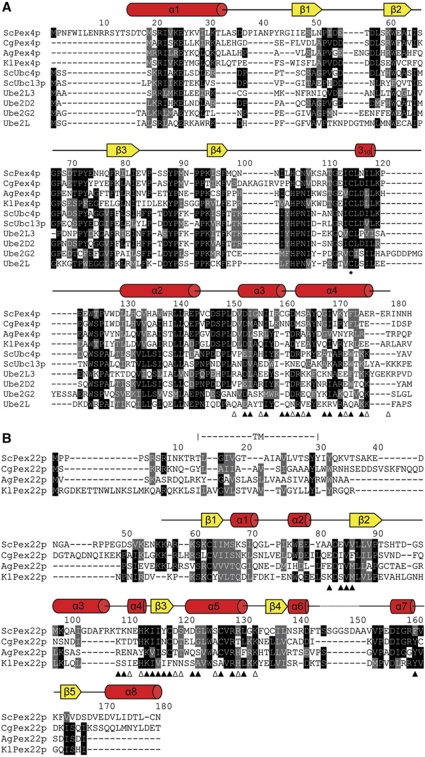
Figure 4.
Sequence alignment of Saccharomyces cerevisiae Pex4p and Pex22p. (A) ScPex4p aligned with a selection of homologous Pex4p sequences and a number of other ubiquitin-conjugating enzymes, allowing comparison with E2s whose functions are not dependent on Pex22p. Secondary structure elements in Pex415–183 are shown above the alignment. The active site cysteine is indicated with an asterisk. Pex415–183 residues involved in interface formation with Pex22S are indicated with triangles below the alignment, with open triangles denoting residues involved in residue-specific polar interactions. Black shading indicates identity and grey shading similarity when present in at least five of the ten sequences aligned. Sc, Saccharomyces cerevisiae; Cg, Candida glabrata; Ag, Ashbya gossypii; Kl, Kluyveromyces lactis. (B) Sequence alignment of Pex22p with homologous sequences. Secondary structure elements, as well as the trans-membrane segment (TM) in ScPex22p, are indicated above the rows. Residues involved in interface formation with Pex415–183 are indicated as in (A). When present in at least three of the four sequences aligned, conservation is indicated as in (A). The four organisms selected in the Pex22p alignment have unambiguously identified Pex4p sequences (A).