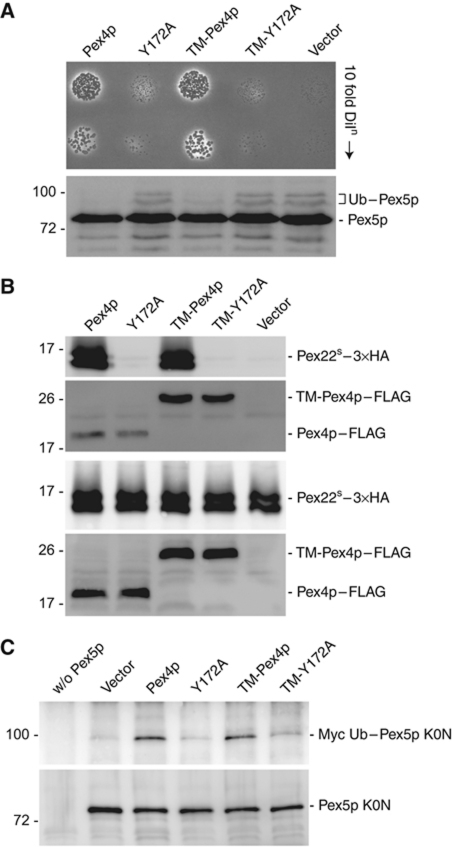
Figure 7.
In vivo analysis of the Pex4p:Pex22p complex. (A) Phenotypical analysis of Pex4p mutants disturbed in Pex22p binding. pex4Δ cells expressing Pex4p variants or an empty vector were grown on media containing oleic acid as sole carbon source for 6 days at 28°C (upper panel) or lysed and samples subjected to SDS–PAGE, western blotting and staining with anti-Pex5p antibodies (lower panel). Ub–Pex5p denotes Ubc4p-modified forms of Pex5p. (B) Pex22p binding capabilities of Pex4p in vivo. Lysates of cells (lower panels) expressing wild-type or mutant forms of Pex4p–FLAG and TM-Pex4p–FLAG, together with 3 × HA-tagged Pex22S were subjected to co-immunoprecipitation with anti-FLAG antibodies and samples were probed with anti-FLAG and anti-HA (upper panels). (C) Ability of Pex4p mutants disturbed in Pex22p binding to modify Pex5p. pex4Δpex5Δ cells, co-expressing Myc-tagged Ub, the Pex5p K0N variant and different Pex4p mutants, were lysed and subjected to immunoprecipitation using Pex5p antibodies. Immunoprecipitates were probed with SDS–PAGE, western blotting and staining with anti-Myc (upper panel) or anti-Pex5p (lower panel) antibodies. Disturbing the interaction between Pex4p and Pex22p (B) results in a growth defect on oleic acid (A) and Ubc4p-dependent ubiquitination of Pex5p (A), a phenotype caused by the inability of Pex4p to ubiquitinate Pex5p on its conserved cysteine residue (C).