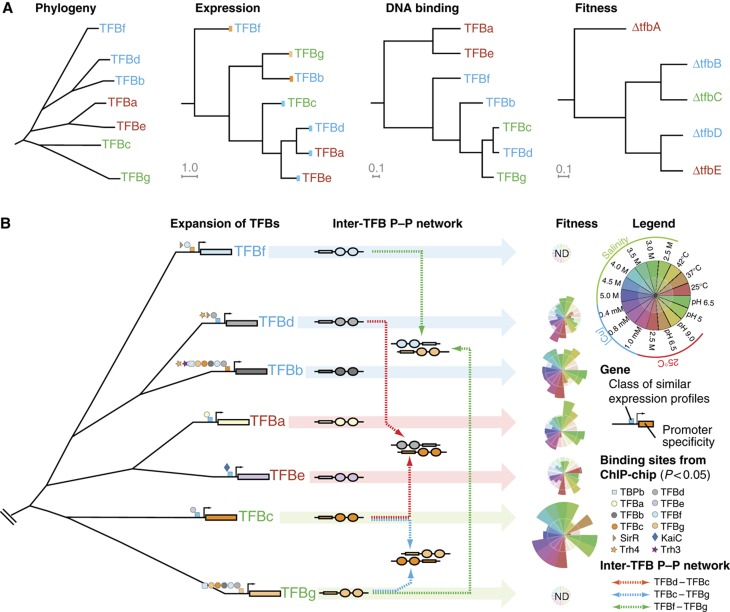
Figure 4.
Reconstruction of evolutionary events responsible for the extant architecture of the seven TFB GRN in H. salinarum NRC-1. (A) Relationships among TFBs at the level of their phylogeny, regulation, distribution of their DNA-binding locations, and fitness contributions. Font coloring of TFBs indicates their clade membership. The first tree shows phylogenetic relationships of TFBs based on the amino-acid sequence similarities. The second tree illustrates relationships in regulation (‘cis-mutations’) of TFBs that were determined by hierarchical clustering of their transcript level changes across 361 environmental conditions. It is clear from this tree that TFBs from the same clade (see b/d/f and g/c clades) are expressed under very different regulatory schemes. The blue and orange color bars on the leaves of this tree indicate related expression profiles; this color code is also utilized in (B) to help the reader relate these data across the two panels. Relationships at the level of DNA binding (‘trans-mutations’) were determined by clustering the hypergeometric P-values for shared-binding sites among pairs of TFBs (Supplementary Table S6). This plot reveals that similarity of DNA-binding specificity is mostly consistent with TFB relationships at the primary sequence level with some important exceptions (see text for details). Finally, similarities in fitness contributions of TFBs across 17 different environments are explained by a combination of cis- and trans-mutations (see text for details). (B) Changes to both cis and trans segments of TFBs need to be considered to explain current day architecture of the seven TFB GRN. This reconstruction was done in the framework of gene duplication events that were inferred from phylogenetic analysis. Promoter evolution was reconstructed by integrating experimentally mapped TF-binding sites (Facciotti et al, 2007) of eight GTFs and four regulators in the TFB promoters, and transcript level changes (A; see inset key). This reconstruction explains subtle differences in the regulation of phylogenetically related TFBs in context of gain and loss of TF-binding sites (for instance, relative to TFBb, the TFBd promoter has gained a TF-binding site for SirR but lost TF-binding sites for six GTFs and Trh3). This reconstruction also reveals convergent evolution of promoters for TFBs from different clades (for instance, TFBc and TFBe); notably, the set of TFs whose TF-binding sites were mapped do not explain the similar expression profiles of TFBc and TFBe. An intra-TFB protein–protein network occurs away from DNA and is speculated to modulate recruitment of these factors to cognate promoters. Coupled changes in DNA-binding specificities of TFBs, their regulation and their protein interactions mediates transcriptional segregation of different aspects of physiology and corresponding environment-specific subfunctionalization of individual TFBs (height of a colored sector in each star plot is proportional to the normalized fitness contribution of that TFB in a particular environment; see inset).