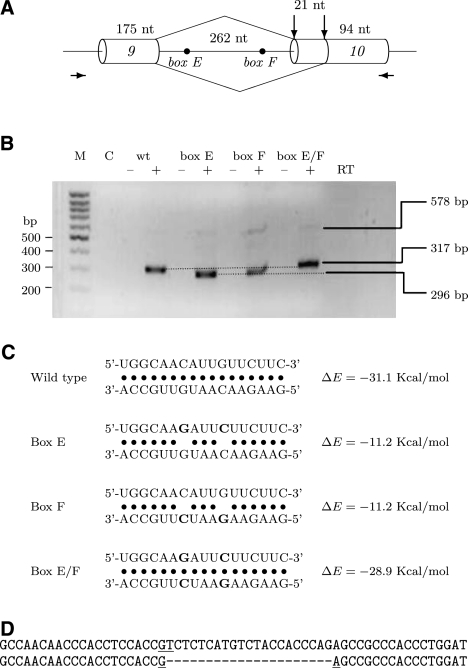
FIGURE 5.
Splicing to alternative acceptor site in the SF1 minigene is regulated by the stem structure formed by the conserved box sequences. (A) Schematic representation of the SF1 minigene, which contains the chromosomal region chr11:64,535,223-64,535,752 (UCSC Genome Browser) from exon 9 to exon 10 of the SF1 gene. Alternative acceptor sites are shown by vertical arrows; locations of primers used for amplification of minigene are indicated by horizontal arrows. (B) Secondary structure formed by the conserved boxes affects acceptor site usage. mRNA products expressed from wild-type (wt) minigene and minigenes mutated within conserved boxes E, F, or both (E/F) were reverse-transcribed and analyzed in 2% agarose gel. (M) size markers (100 bp DNA ladder), (C) control (PCR in the absence of template). The addition (+) or absence (−) of the reverse transcriptase (RT) enzyme to the reaction is indicated. The positions of unspliced (578-bp) and spliced (317- bp and 296-bp) products are shown on the right. (C) Predicted base-pairing for the wild type, box E, box F, and box E/F mutants (point mutations are shown in boldface), with the estimated equilibrium free energies. The box E sequence is shown above the box F sequence. (D) Comparing the nucleotide sequences of alternatively spliced products of minigenes: wild type and box E/F mutant (upper) and box E and box F mutant (lower).