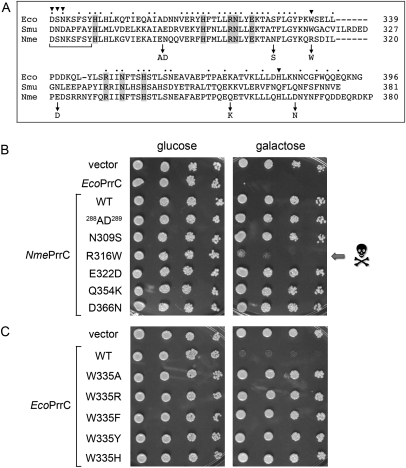
FIGURE 3.
A gain-of-function mutation renders NmePrrC toxic to yeast. (A) The amino acid sequence of the nuclease domain of EcoPrrC is aligned to the homologous segments of SmuPrrC and NmePrrC. Positions of side-chain identity/similarity in all three proteins are indicated by • above the alignment. The eight conserved residues defined previously as essential for yeast toxicity are shaded gray. Positions of side chain variation between the nontoxic NmePrrC and the toxic Eco and Smu PrrCs are indicated by arrows below the alignment, which specify the NmePrrC mutations tested for gain-of-toxicity in yeast. The LARP motif is demarcated by the bracket below the sequences. EcoPrrC residues subjected to mutational analysis in the present study are indicated by ▾. (B) Serial fivefold dilutions of yeast cells bearing a CEN plasmid encoding the indicated galactose-regulated prrC gene or an empty CEN vector were spotted on –Leu agar plates containing glucose or galactose as specified. (C) Toxicity tests for wild-type EcoPrrC and the indicated W335 mutants are shown.