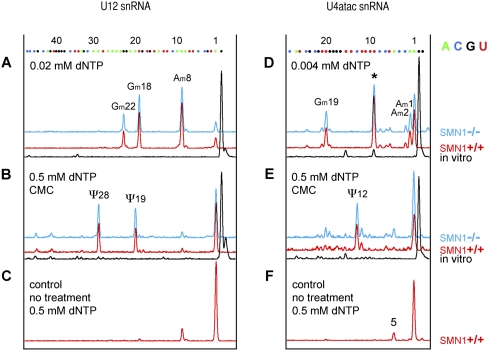
FIGURE 3.
Mapping of modified nucleotides in minor snRNAs U12 (left) and U4atac (right). (A) Three 2′-O-methyl groups are detectable in U12 snRNA from SMN1+/+ (red) and SMN1−/− (blue) lymphoblasts (Am8, Gm18, Gm22). (B) Two pseudouridines are found in U12 snRNA from SMN1+/+ (red) and SMN1−/− (blue) lymphoblasts (Ψ19 and Ψ28) and no peak is observed for in vitro-transcribed U12 snRNA (black). (C) Only a tiny peak for Am8 is detectable in control untreated RNA. (D) 2′-O-methylation is detectable in U4atac at positions A1, A2, an G19. A prominent stop is observed also at position 9 (star) suggesting a 2′-O-methyl group at C8, but the reaction is terminated at the same position when in vitro-transcribed RNA is used (black trace). (E) A pseudouridine at position 12 is detectable in U4atac from SMN1+/+ (red) and SMN1−/− (blue) lymphoblasts, but not in control in vitro-transcribed U4atac (black). (F) When control primer extension reactions are run on RNA samples isolated from lymphoblasts an extra peak appears at position 5. This might correspond to a different isoform of human U4atac. In fact, 19 different genes are reported for U4atac in the human genome databases (http://useast.ensembl.org/Homo_sapiens/) and the U4atac.14-201 variant fits the detected size.