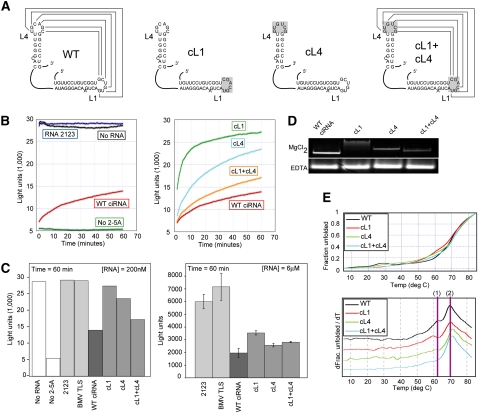
FIGURE 2.
Function and structural role of the kissing hairpins. (A) Diagram of the mutants used to study the structural role of the kissing hairpin interaction. For clarity, only the L1 and L4 elements are shown. The loop(s) mutated in each RNA are shaded gray. (B, left) Time course of fluorescence from a representative in vitro RNase L cleavage assay. Controls with no added RNA, with a negative control RNA (2123), or without 2-5A activator are shown. The inhibition effect of WT ciRNA also is included. (Right) Comparison of WT, cL1, cL4, and cL1+cL4 mutant RNAs in the cleavage assay. All concentrations of RNA = 200 nM. (C, left) Bar graph depiction of the experiment shown in panel B. The amount of fluorescence at time = 60 min is shown for each experiment. An additional negative control RNA (BMV TLS, a structured RNA form the 3′ UTR of the brome mosaic virus genomic RNA) also is included. One representative experiment is shown. (Right) Bar graph of an in vitro cleavage assay done at elevated RNA concentrations (6 μM) showing how the mutants inhibit nearly as well as WT at this concentration of RNA. Data are three averaged experiments; error bars, 1 SD from the mean. (D) Native gels of WT ciRNA and the cL1, cL4, and cL1+cL4 mutants. The top gel contained 5 mM MgCl2; the bottom contained 2 mM EDTA. (E) Thermal denaturing curves of WT ciRNA and the three kissing loop mutants. At top is the raw data normalized to reflect fraction RNA folded as a function of temperature; at bottom is the first derivative of the data. The two transitions discussed in the text are indicated.