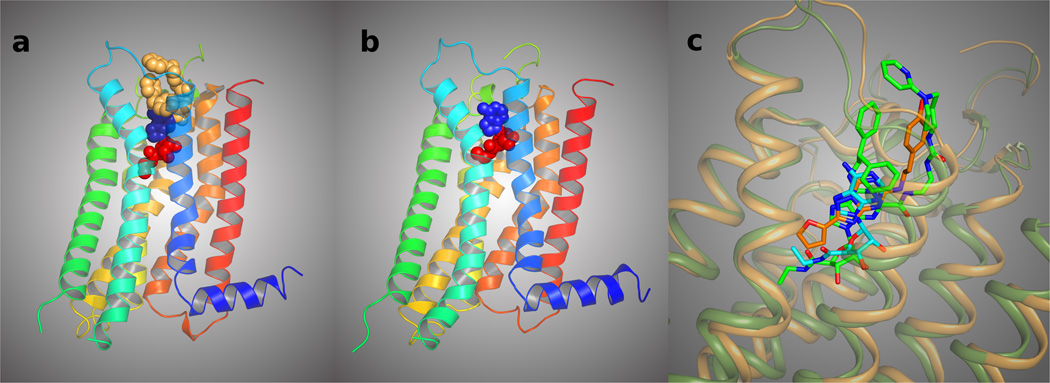
Figure 1.
A. The newly determined structure of the A2AAR is shown surrounding its synthetic agonist 1.16 The helices (color sequence from red to blue) and the slim connecting loops represent the receptor protein, winding back and forth through the cell membrane. The central ribose moiety (red) of the agonist binds in a hydrophilic region and is critical for activation of the receptor, while the adenine heterocycle (blue) binds in a hydrophobic region. The top (tan-colored) C2 and N6 substituents of the agonist, facing the outside of the cell, effectively fill the remaining spaces in the binding site and stabilize the receptor in order to obtain a crystallized structure. B. Similar view of the agonist docking model of Ivanov et al. using the inactive A2AAR structure.4,19 The potent nonselective agonist 14 is present in the binding site. C. Superposition of the agonist 1 (carbons colored in green) in 1-A2AAR (represented as ribbon colored in green), the antagonist 2 (carbons colored in orange) in 2-A2AAR (represented as ribbon colored in orange), and the predicted docked pose of 14 as in Ivanov et al. (carbons colored in cyan).