Figure 1.
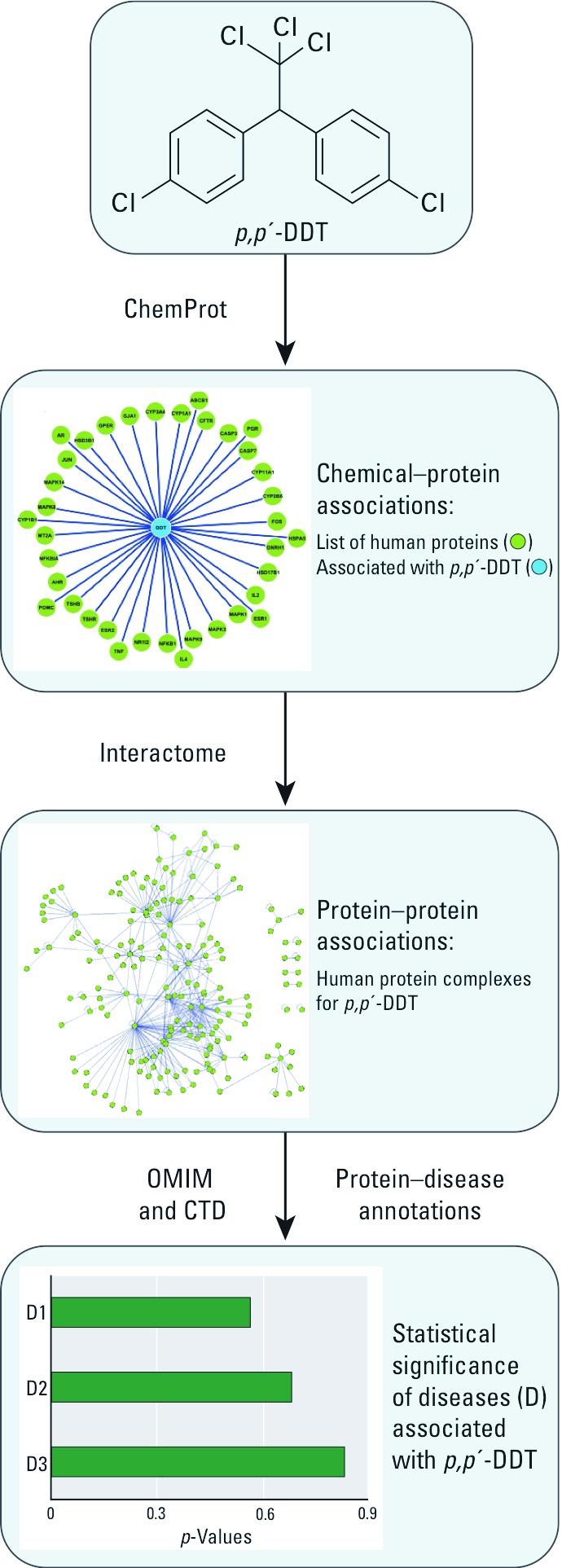
Overview of the systems chemical biology three-step approach. (1) Extraction of existing knowledge using a disease chemical biology database (ChemProt) to generate chemical–protein networks for p,p´-DDT. (2) Creation of protein complexes by protein enrichment using a high-confidence set of experimental protein–protein interactions. (3) Statistical ranking of diseases (D) known or predicted to be linked to p,p´-DDT after integration of protein–disease annotations to protein complexes based on information in the Online Mendelian Inheritance in Man (OMIM) database and the Comparative Toxicogenomics Database (CTD).
