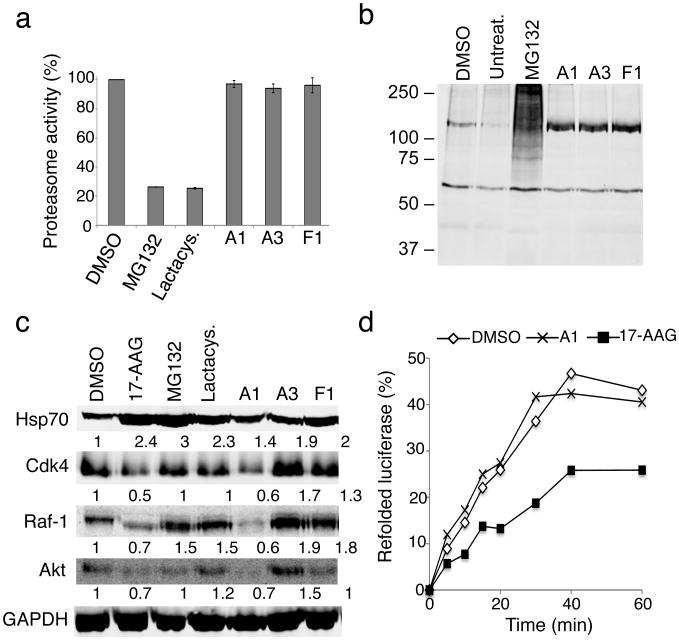
Figure 7. The PRs are not proteasome or Hsp90 inhibitors.
(a) HeLa cells were incubated with either DMSO, MG132 (10 μM), lactacystin (lactacys., 6 μM) and the PRs A1, A3 and F1 (10 μM) for 6 h. Proteasome-associated chymotrypsin activity was assessed using the fluorogenic substrate Suc-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (suc-LLVY-AMC) as described in Materials and Methods. (b) HeLa cells were either left untreated or treated with DMSO, the proteasome inhibitor MG132 (10 μM), or the PRs A1, A3 and F1 (10 μM) for 16 h. Whole cell extracts of HeLa cells were separated by SDS-PAGE, transferred to membranes, stained with Ponceau S to visualize total protein and probed using a rabbit polyclonal antibody to detect ubiquitin. (c) HeLa cells were treated with either DMSO, 17-AAG (2 μM), MG132 (10 μM), lactacystin (lactacys., 6 μM) or the PRs A1, A3 and F1 (10 μM) for 24 hr. Protein levels of various Hsp90 client proteins (Cdk-4, Raf-1 and Akt) in equal amounts of whole-cell lysates were assessed by western blot analysis. GAPDH was used as loading control. Densitometric measurements of Hsp90 client protein levels normalized to GAPDH in relation to control DMSO-treated cells were performed using ImageJ software. (d) Refolding of chemically-denatured firefly luciferase was assessed in RRL containing 2 mM ATP in the presence of either DMSO (○), 17-AAG (2 μM, ■) or the PR A1 (10 μM, ×). Luciferase activities are expressed as percent of the native enzyme control. The result shown is representative of three experiments.