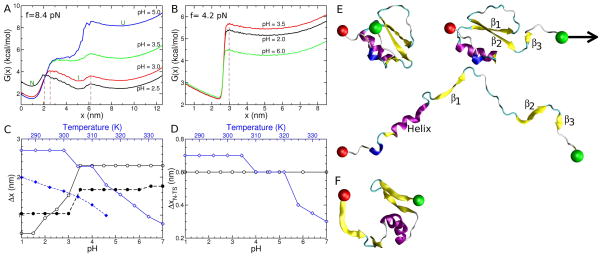
Figure 5.
pH and temperature effects on the mechanical response of CI2 and protein G at a constant tension force of 8.4 pN and 4.2 pN, respectively. The free energy profile G(x) = −kBTln(P(x)), where P(x) is the probability of finding a given x value, as a function of the end-to-end distance of the protein, projected onto the pulling vector, for (A) CI2 and (B) protein G at different pH values as labeled. The temperature is 302 K and 317 K in (A) and (B), respectively. Brown dashed lines indicate transition state locations at pH=2.5 and 3.0 in (A), and pH=3.5 in (B). The location of the native, intermediate, and fully unfolded basins of attraction in G(x) are marked by the labels N, I, and U, respectively. (C) For CI2, the distance, Δx, between the native and first transition state (ΔxN–TS), and intermediate and second transition state (ΔxI–TS) are shown as a function of pH (lower axis) and temperature (upper axis). The black symbols correspond to pH and blue symbols are for temperature. In both cases solid lines show ΔxN–TS and dashed lines correspond to ΔxI–TS. (D) Same as (C) but for protein G. No intermediate basin of attraction exists for protein G, so only the distance between the native and transition state (ΔxN–TS) is reported. (E) Sample conformations (top to bottom) from the native, intermediate and unfolded states of CI2 during simulations at 300 K and f = 8.68 pN. β-strands 1 through 3 are labeled and the direction in which the constant tension is applied to the C-terminus (green sphere) is indicated by the black arrow. The N-terminal residue, fixed in space during the simulation is shown as a red sphere. (F) Simulation structure of protein G with x=3.09 nm from the replica at T=320 K and f = 4.1 pN in the replica exchange simulations.