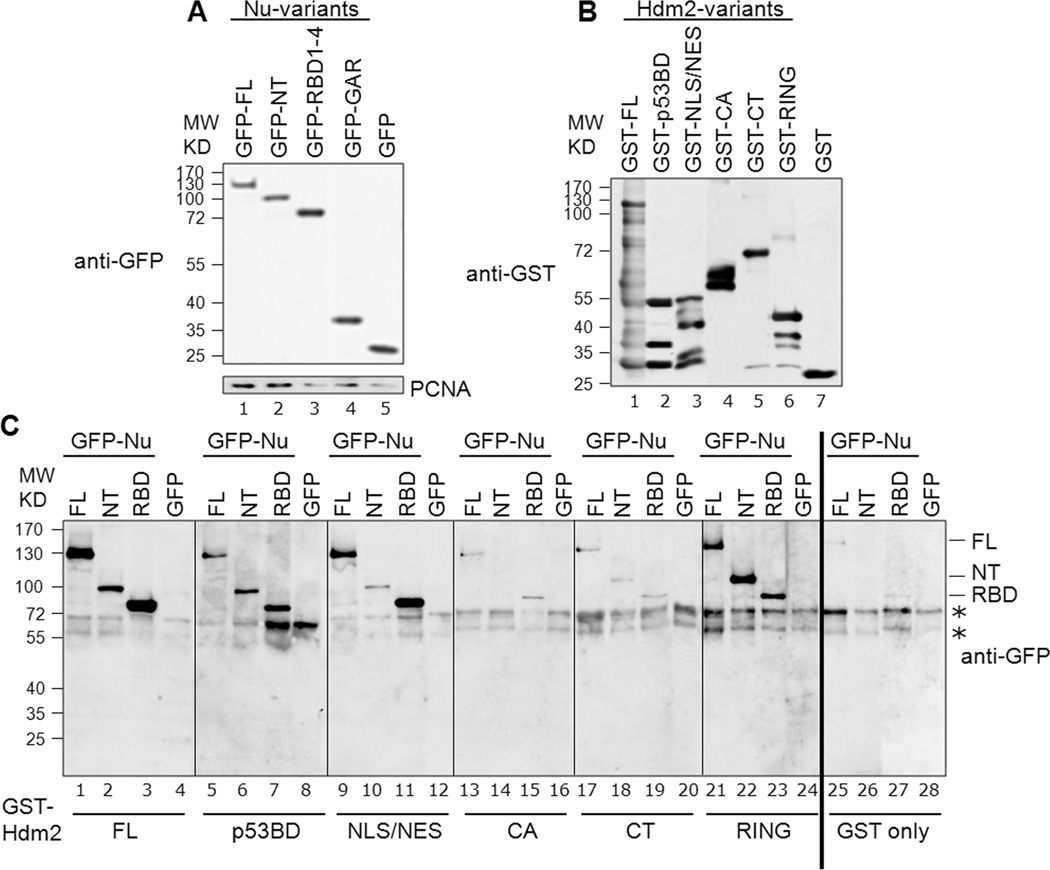
Fig. 3. Determination of specific domains involved in nucleolin-Hdm2 interactions using cell lysates.
A, Representative expression of the GFP-tagged nucleolin constructs: The various nucleolin constructs, as indicated, were transfected into H1299 cells. Lysates were prepared from these cells and examined by Western blotting for GFP. PCNA served as a loading control. B, Representative corresponding GST-tagged Hdm2 variants utilized in a given binding reaction: For a given binding reaction, the same blot was analyzed following stripping of anti-GFP and reprobing with anti-GST. Only one representative lane for each is shown and rearranged for easier display. C, Identification of interacting nucleolin and Hdm2 domains. Binding assays were performed using equivalent amounts of GST-tagged Hdm2 variants bound to glutathione-Sepharose beads. These constructs were Hdm2 FL (lanes 1–4), p53-BD (lanes 5–8), NLS/NES (lanes 9–12), CA (lanes 13–16), CT (lanes 17–20) and RING (lanes 21–24). GST alone was used as the negative control (lanes 25–28). The bead-bound Hdm2 variants were incubated with 1 mg of protein lysates from H1299 cells expressing GFP-tagged nucleolin FL, NT, RBD1–4 (RBD), or GFP alone, as indicated. The presence of nucleolin (full-length and truncation domains) was determined by subjecting the bead-bound material to Western analysis, probing for GFP. The non-specific signals seen in the GFP blot are indicated by asterisks. All the blots for this figure were exposed to film simultaneously and developed for identical time periods. After scanning, the particular sections were cropped and rearranged for easier display.