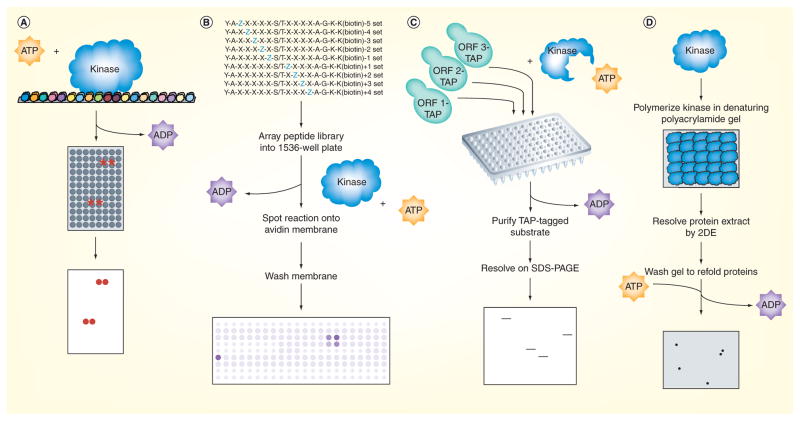
Figure 1. Phosphorylation mapping on a global scale.
(A) Protein microarrays containing approximately 4400 of the approximately 6000 yeast proteins spotted in duplicate at high spatial density onto glass slides were incubated with kinase and radiolabeled ATP, washed and developed by autoradiography [4]. Positive substrates were determined by quantifying the amount of radiolabel incorporated at each pair of spots relative to the corresponding pair on a control slide assayed in parallel in the absence of kinase. (B) The peptide library used by Mok et al. was a positional-scanning solution-phase library made up of 198 distinct mixtures of biotinylated 16-mers, which each had a central serine or threonine residue as a phosphoacceptor site, and a different amino acid residue fixed at one of the nine positions surrounding the phosphoacceptor site [15]. Kinases were assayed against the peptides in 1536-well plates using radiolabeled ATP, and upon completion of the reaction, the peptides were spotted onto an avidin-impregnated membrane that was then washed and exposed to a phosphoimager. The extent of phosphorylation of each mixture was quantified to generate a motif representing the amino acid preferences targeted by the kinase. (C) Analog-sensitive kinase alleles were generated by mutating the ATP binding pocket; such binding is favored for an ATP analog that cannot be accommodated by wild-type kinases. Lysates prepared individually from strains containing candidate substrates epitope-tagged at their endogenous locus were mixed with the purified kinase and a radiolabeled form of the ATP analog. The candidate substrates were then purified and resolved by gel electrophoresis before being exposed by autoradiography. Positive substrates were determined as those bands appearing on the autoradiograph indicating incorporation of the radiolabeled phosphate. (D) In reverse in-gel kinase assay, the kinase of interest was first polymerized in a denaturing polyacrylamide gel that was subsequently used to resolve a protein extract containing the candidate substrates by 2DE. The gel was then washed thoroughly to remove the SDS detergent and refold the proteins into their native forms, incubated in kinase buffer containing radiolabeled ATP and exposed using autoradiography. Positive substrates were determined as those spots appearing on the radiograph having incorporated the radiolabeled phosphate. The identities of the novel substrates were determined by excising the corresponding spots from a silver-stained gel run in parallel without any kinase and analyzing them by mass spectrometry.
ORF: Open reading frame; TAP: Tandem affinity purification.