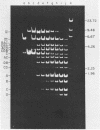
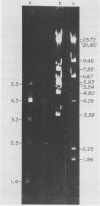
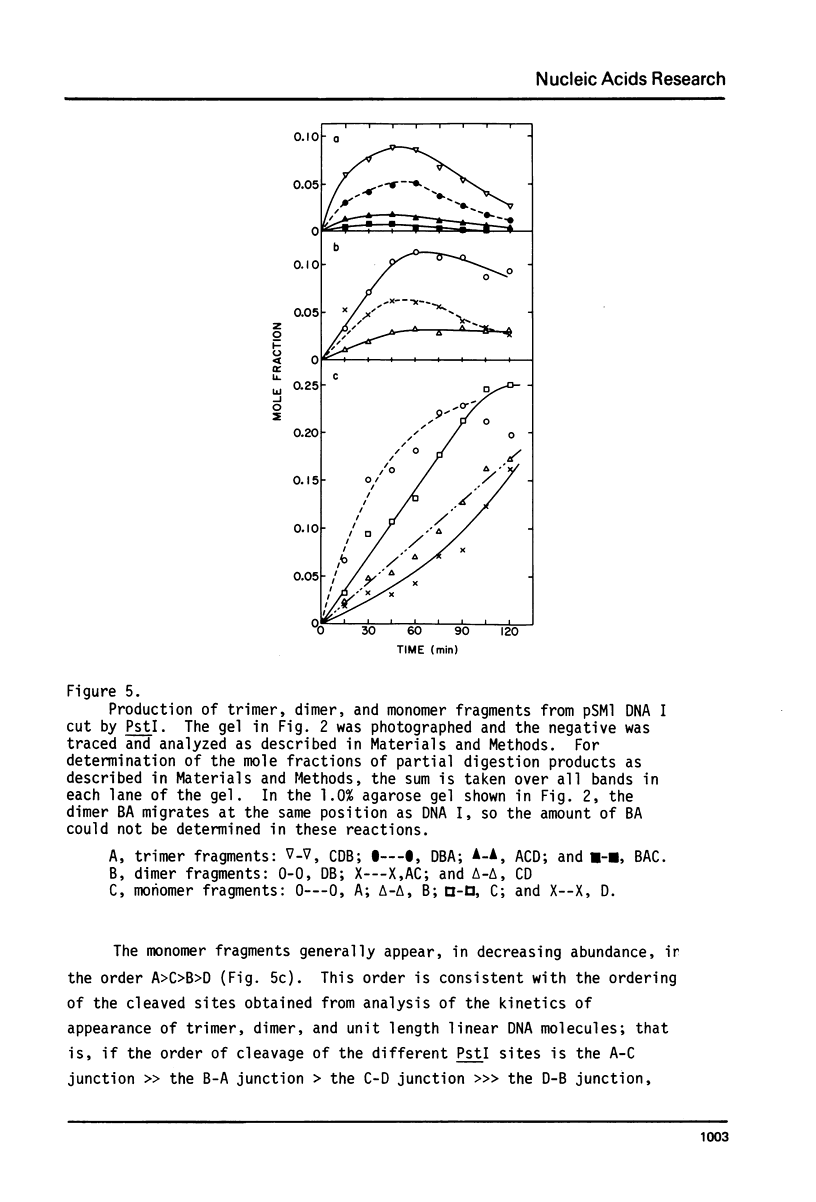
Abstract
The four identical recognition sites for the restriction endonuclease PstI in purified plasmid pSM1 DNA I are cleaved at markedly different rates. The order and relative frequencies of cleavage at these four PstI sites have been determined from the order of appearance of partial cleavage products and from an analysis of production of specific unit length linear molecules. The same pattern of preferential cleavage is also found when linear, nicked circular, or relaxed closed circular forms of the same plasmid DNa are used as substrates for PstI. Inspection of the nucleotide sequences immediately adjoining each of the PstI sites suggests that the presence of adjacent runs of G-C base pairs confers significant resistance to cleavage.
Full text
PDF


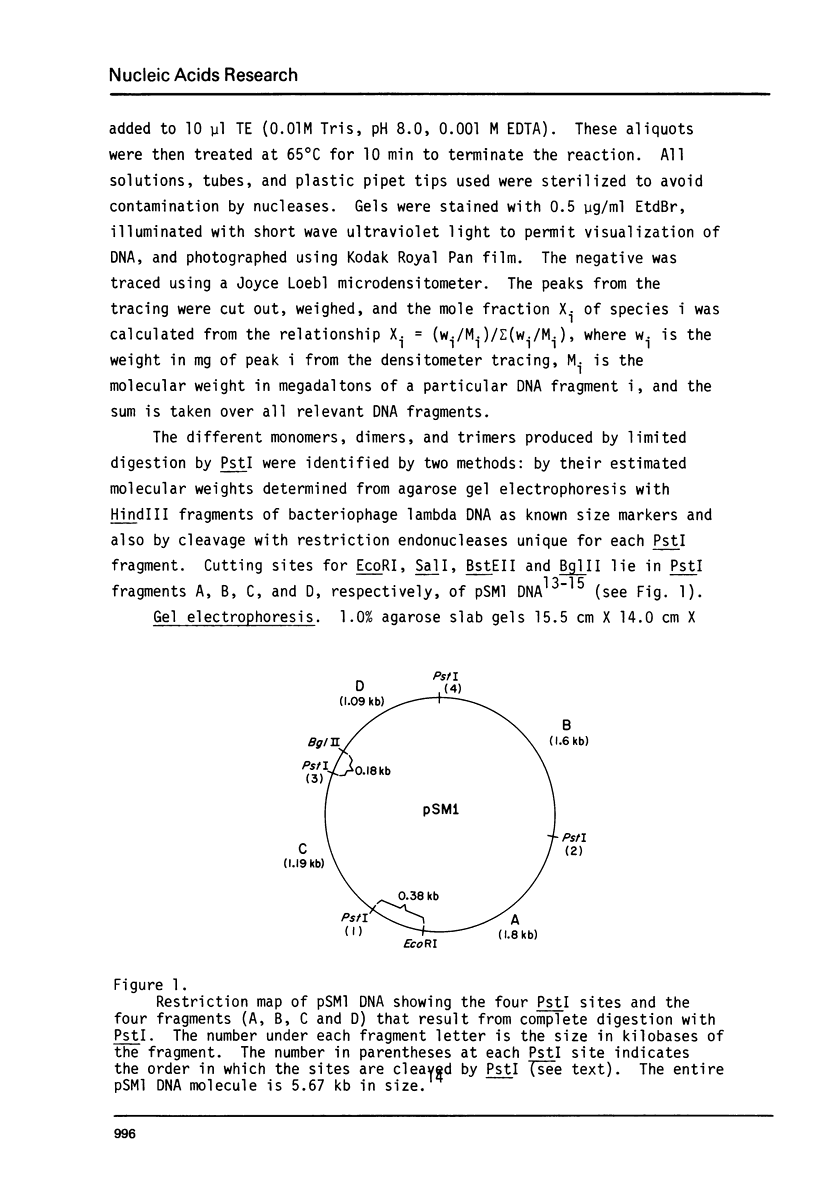



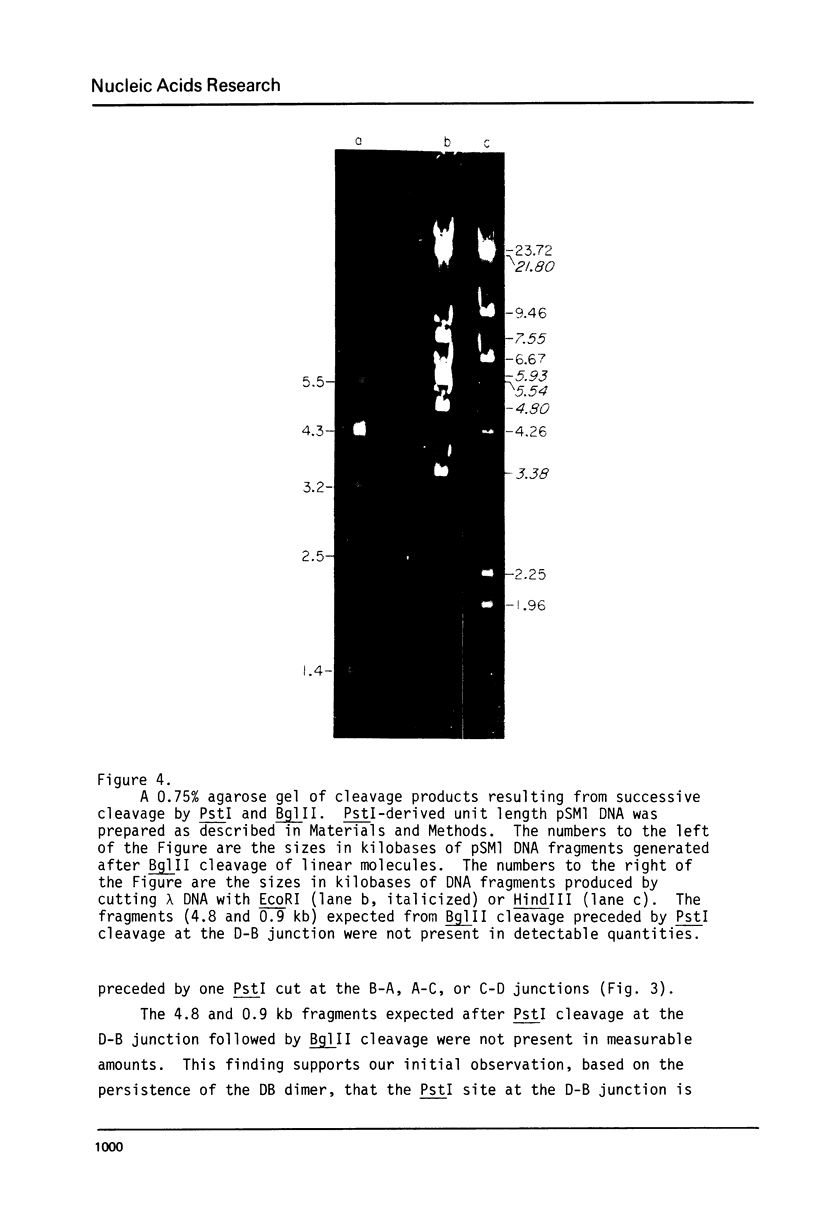






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Smith M. The mapping and sequence determination of the single site in phiX174am3 replicative form DNA cleaved by restriction endonuclease Pst I. FEBS Lett. 1976 Jun 15;65(3):284–287. doi: 10.1016/0014-5793(76)80130-5. [DOI] [PubMed] [Google Scholar]
- Burke R. L., Bauer W. Measurement of superhelix densities in buoyant dye/CsCl. The use of a standard other than native SV40 DNA. J Biol Chem. 1977 Jan 10;252(1):291–292. [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Drew H., Takano T., Tanaka S., Itakura K., Dickerson R. E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980 Aug 7;286(5773):567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- Forsblom S., Rigler R., Ehrenberg M., Philipson L. Kinetic studies on the cleavage of adenovirus DNA by restriction endonuclease Eco RI. Nucleic Acids Res. 1976 Dec;3(12):3255–3269. doi: 10.1093/nar/3.12.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L., Thomas M., Davis R. W. EcoRI endonuclease cleavage map of bacteriophage P4-DNA. Virology. 1975 Aug;66(2):420–427. doi: 10.1016/0042-6822(75)90214-7. [DOI] [PubMed] [Google Scholar]
- Kahn M., Kolter R., Thomas C., Figurski D., Meyer R., Remaut E., Helinski D. R. Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol. 1979;68:268–280. doi: 10.1016/0076-6879(79)68019-9. [DOI] [PubMed] [Google Scholar]
- Mickel S., Bauer W. Isolation, by tetracycline selection, of small plasmids derived from R-factor R12 in Escherichia coli K-12. J Bacteriol. 1976 Jul;127(1):644–655. doi: 10.1128/jb.127.1.644-655.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickel S., Ohtsubo E., Bauer W. Heteroduplex mapping of small plasmids derived from R-factor R12: in vivo recombination occurs at IS1 insertion sequences. Gene. 1977;2(3-4):193–210. doi: 10.1016/0378-1119(77)90017-8. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Feingold J., Ohtsubo H., Mickel S., Bauer W. Unidirectional replication in Escherichia coli of three small plasmids derived from R factor R12. Plasmid. 1977 Nov;1(1):8–18. doi: 10.1016/0147-619x(77)90004-x. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H., McCormick M., Machida C., Machida Y. Mechanism of insertion and cointegration mediated by IS1 and Tn3. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):283–295. doi: 10.1101/sqb.1981.045.01.041. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H. Plasmids containing insertion elements are potential transposons. Proc Natl Acad Sci U S A. 1980 Feb;77(2):750–754. doi: 10.1073/pnas.77.2.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka A. Recovery of DNA fragments inserted by the "tailing" method: regeneration of PstI restriction sites. Gene. 1981 May;13(4):339–346. doi: 10.1016/0378-1119(81)90013-5. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Two restriction endonucleases from Bacillus globiggi. Nucleic Acids Res. 1976 Jul;3(7):1747–1760. doi: 10.1093/nar/3.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullman B., Pullman A. Quantum-mechanical investigations of the electronic structure of nucleic acids and their constituents. Prog Nucleic Acid Res Mol Biol. 1969;9:327–402. doi: 10.1016/s0079-6603(08)60772-2. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Inokuchi H., Ohtsubo H., Ohtsubo E. Genes and sites involved in replication and incompatibility of an R100 plasmid derivative based on nucleotide sequence analysis. Mol Gen Genet. 1980;179(3):527–537. doi: 10.1007/BF00271742. [DOI] [PubMed] [Google Scholar]
- Rubin R. A., Modrich P. Substrate dependence of the mechanism of EcoRI endonuclease. Nucleic Acids Res. 1978 Aug;5(8):2991–2997. doi: 10.1093/nar/5.8.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. I., Blattner F. R., Davies J. The isolation and partial characterization of a new restriction endonuclease from Providencia stuartii. Nucleic Acids Res. 1976 Feb;3(2):343–353. doi: 10.1093/nar/3.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]