Fig. 5. Structural changes in the AphB N100E variant.
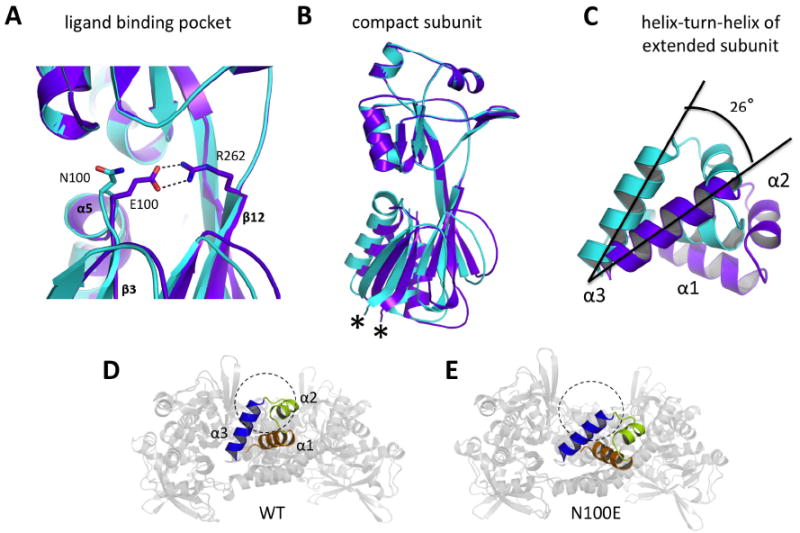
(A) Formation of a salt bridge between E100 and R262 in the regulatory domain of N100E moves the β3-α5 loop 1.3 Å closer to β12. Wild-type AphB, cyan; N100E, purple. (B) The RD-I of N100E moves by up to 3Å at the attachment point of the linker helix (denoted by *) and this movement is relayed to the entire N-terminal domain dimer, which moves as a rigid body. Colors are the same as in (A). (C) Rigid body movement of the N-terminal domain dimers causes a displacement of the DNA-binding domains. The α3 helices of the extended subunits of wild-type and N100E align on their C-terminal ends and the entire helix-turn-helix motif of N100E aligns to that of wild-type upon a 26° counterclockwise rotation. This angle was determined manually, by orienting the WT and N100E recognition helices in MacPymol such that they were on XY plane and measuring the angle between them with a protractor. These results were verified by rotating the N100E helix-turn-helix about the Z-axis (centered on the C-terminal ends of the recognition helices) by -26° in MacPymol. Colors are the same as in (A). (D) View down the DNA-binding groove of the wild-type AphB tetramer showing outer recognition helix (α3) (blue) and the outer α2 helix (lime green) would preclude DNA-binding (DNA cross-section shown as dashed circle) due to steric clashes. (E) View down the DNA-binding groove of the N100E AphB tetramer showing the outer α3 helix (blue) is now poised to interact with the major groove of DNA (DNA cross-section shown as dashed circle) and the α2 helix (lime green) has moved to a position where steric clashes with DNA will not occur.
