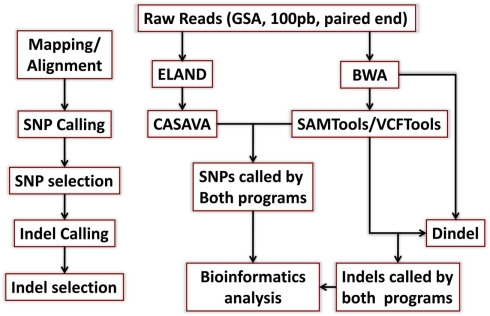
Figure A1.
Schematic of the mapping/alignment and variant calling steps. We used the Eland and BWA aligners to map reads to the reference genome, and CASAVA and SAMTools to call SNPs as explained in Section “Materials and Methods.” Short insertion and deletions were called using SAMTools and Dindel. Only variants that were called by both algorithms and passed quality control filters were included in the follow-up analyses.