Abstract
We present the development of a tool, which provides users with the ability to visualize and interact with a comprehensive description of a multi-scale model of the renal nephron. A one-dimensional anatomical model of the nephron has been created and is used for visualization and modelling of tubule transport in various nephron anatomical segments. Mathematical models of nephron segments are embedded in the one-dimensional model. At the cellular level, these segment models use models encoded in CellML to describe cellular and subcellular transport kinetics. A web-based presentation environment has been developed that allows the user to visualize and navigate through the multi-scale nephron model, including simulation results, at the different spatial scales encompassed by the model description. The Zinc extension to Firefox is used to provide an interactive three-dimensional view of the tubule model and the native Firefox rendering of scalable vector graphics is used to present schematic diagrams for cellular and subcellular scale models. The model viewer is embedded in a web page that dynamically presents content based on user input. For example, when viewing the whole nephron model, the user might be presented with information on the various embedded segment models as they select them in the three-dimensional model view. Alternatively, the user chooses to focus the model viewer on a cellular model located in a particular nephron segment in order to view the various membrane transport proteins. Selecting a specific protein may then present the user with a description of the mathematical model governing the behaviour of that protein—including the mathematical model itself and various simulation experiments used to validate the model against the literature.
Keywords: physiome project, CellML, computational physiology
1. Introduction
Kidneys are vital organs of humans and play an integral part in overall homeostasis of the body. Each kidney is composed of one million nephrons, which are the functional units of the kidney. The kidneys receive approximately 25 per cent of the cardiac output per minute and one of the many functions of this organ is the filtration of blood and the formation of the filtrate that will ultimately become urine. The renal processes of reabsorption and secretion modify the composition (water, ions and solute) of the filtrate as it passes along the length of the nephron.
A nephron is a tubular structure composed of a single layer of epithelial cells (figure 1a) lining a series of segments: proximal tubule (PT), thin descending limb of the loop of Henle, thin and thick ascending limbs of the loop of Henle, the distal tubule and the collecting duct. Each segment of the nephron has specific functions in terms of reabsorption and secretion, and the specificity of function is dependent upon the transport proteins that reside in the apical (lumen facing) and the basolateral membranes (serosal facing, figure 1a).
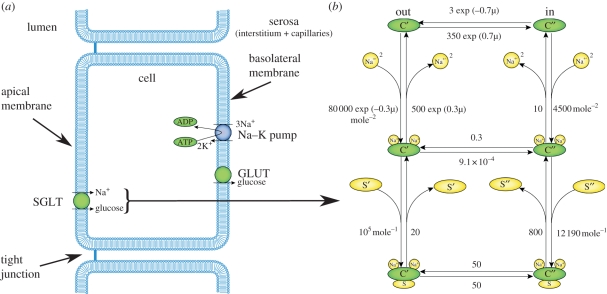
Figure 1.
(a) Schematic diagram illustrating the structure of an epithelial cell of a renal tubule including three of the transport proteins found in the proximal tubule (PT), the sodium–glucose cotransporter (SGLT) located on the apical membrane and the sodium–potassium pump (Na–K pump) and facilitated glucose transporter (GLUT) located on the basolateral membrane. (b) The state transition diagram for the Eskandari et al. [1] SGLT1 model, a specific instance of a model representing the generic transport mechanism shown in (a).
Epithelial cells are polarized cells in which different transport proteins reside within the apical and basolateral membranes (figure 1a). It is this discrete localization of transport proteins which allows the specific reabsorption of a solute across the epithelial cell and, ultimately, its return to the blood. For instance, the reabsorption of sodium from the filtrate of the lumen of the PT back to the blood requires the concerted action of both apical-specific and basolateral-specific transporters. The sodium–potassium pump (Na–K pump) is a primary active transport protein located within the basolateral membrane of a cell and uses ATP to transport sodium against its electrochemical gradient, from the cell into the interstitium (from whence it can diffuse back into the blood; figure 1a). The action of the Na–K pump lowers the concentration of sodium inside the cell and thus establishes the electrochemical gradient favouring sodium entry from the lumen of the tubule into the cell.
There are many sodium-dependent cotransporters (symporters) and counter-transporters (antiporters) that use the sodium electrochemical gradient to ‘power’ the ‘uphill’ transport of other ions and solutes (against their electrochemical gradient). For example, the sodium–glucose cotransporters (SGLTs) are secondary active transporters which couple the uptake of glucose up its concentration gradient from the tubule lumen of the PT to the transport of sodium down its electrochemical gradient (figure 1, [2]), thus concentrating glucose in the cell. The second step in the process of glucose reabsorption is the transport of glucose (figure 1a), into the interstitial space and back into the circulation. This occurs by means of a facilitated glucose uniporter (GLUT family) that transports glucose down the concentration gradient across the basolateral membrane [3]. In a mathematical model of the sodium–glucose cotransporter (SGLT1), Wright and colleagues [1,4] have used a six-state model to represent the kinetics of the individual steps in the transport cycle (figure 1b). The binding of drugs to a specific state of the cycle can be modelled [4], and follow-on effects (on whole-cell and tubule function) can be simulated.
Many transport proteins have the potential of being targets for the treatment of disease. Indeed, SGLT2 has tremendous promise as a target for the possible treatment of hyperglycaemia and obesity in patients with type 1 or type 2 diabetes [5]. However, the effect of a drug must be assessed in non-human systems (e.g. laboratory animal testing), which is very expensive, prior to human trials. Alternative strategies must be made available. The field of mathematical modelling of physiology has this potential. Modelling the function of transport proteins is an innovative diagnostic tool in which testing of therapeutic drugs (e.g. effects of dapagliflozin on the function of SGLT2) can be assessed on altering transport protein function. Of course, the first step in this endeavour is to create an interactive mathematical model of the nephron.
In addition to the potential use of mathematical models of the nephron in a clinical application, these models have potential importance as teaching tools. In this day and age of multi-media, educational technology, a physiology teacher's tools now not only encompass standard didactic lectures and associated practical laboratories, but also self-directed learning (e.g. problem-based case studies), Internet-based distance learning and emerging (web-based) mathematical modelling of systems physiology (e.g. Web-HUMAN, http://placid.skidmore.edu/human/). For instance, when teaching renal physiology, one of the initial concepts introduced to the students is the basic structure of the nephron and to explain the inter-relationship of many nephrons working in unison for proper function of the kidney. Explaining the structure of the nephron might adequately be performed by a two-dimensional PowerPoint presentation; however, a structure (e.g. a series of nephrons) with a three-dimensional architecture is more challenging for the students to visualize. Thus, multi-scale modelling of the nephron(s) provides a three-dimensional interactive tool, which students can use in their efforts of understanding the overall function of a multi-nephron system.
Multi-scale mathematical models of a nephron incorporate biophysical detail from the level of individual transport proteins to the flow of fluid through the lumen of the whole nephron tubule. Owing to the complexity of these multi-scale models, it is difficult to communicate the details of the model structure, and the results, by means of a static description of the model. We present here the development of a tool to facilitate the presentation and interrogation of multi-scale nephron models. This prototype interface provides the user with an anatomical browser, cellular and subcellular transport model database, and some preliminary simulation results. Furthermore, the models of the cells and the transport proteins are encoded in the model description format CellML ([6], http://www.cellml.org). This approach facilitates and enhances the ability of the modellers to share and collaborate with other scientists.
2. Methods
We have developed a user interface that allows the visualization and interaction with the multi-scale nephron models and simulation results. The user interface has been developed as a web-delivered application viewable from a standard web browser. This tool is being designed in conjunction with mathematical model description and annotation methods and tools, thus we aim to automatically generate the content of this tool directly from the annotated model descriptions. In this manner, the actual presentation of the content to the user is able to be customized to suit specific user requirements. In the following, we describe the framework we have developed in the context of a specific prototype presentation configuration. It is important, however, to remember that this is just one possible presentation configuration of the underlying data. We will endeavour to highlight the ability of our framework to enable different configurations as appropriate in the following sections.
The basis of the framework we are developing is a comprehensive and descriptive anatomical and physiological model of the renal nephron. This includes geometric models and mathematical models of renal function spanning multiple spatial scales. Previously, we have presented methods for capturing such comprehensive descriptions for models that can be encoded in CellML [7,8]. Current language specifications and tool support for CellML enable the encoding of a large range of cellular and subcellular processes and their assembly into whole cell models [9,10]. Larger spatial scale-lumped parameter models are also able to be encoded in CellML [11], but spatially distributed models are not. FieldML [12] has been proposed as a common method for encoding such models and we have previously explored how FieldML models can be annotated in a similar manner to CellML models and integrated into our concept of a comprehensive description [13].
In the current work, we endeavour to use commonly used formats where appropriate and sufficient tool support is available. However, we are restricted to using custom software to implement certain aspects of the renal nephron we present here until the FieldML language specification stabilizes and tool support improves. The consequence of this is the need to hand-craft portions of the content data we wish to present in our user interface. By ensuring we follow the methods developed previously [7,8,10,13] when authoring these hand-crafted portions of the content we expect to be able to plug in future formats, such as FieldML, as they become available.
2.1. Tool design
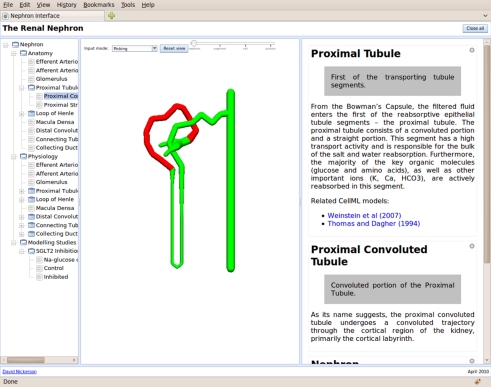
The prototype user interface presentation consists of three primary regions: the content tree, a graphical view and the information panel (figure 2). The content tree provides a primary overview of the content of the interface. The graphical view panel provides space for graphical presentation and interaction. The information panel displays detailed information in response to user input via the content tree, graphical view or information panel. In figure 2, the PT component can be seen (matching the current selection in the content tree and graphical view) consisting of a brief description of the component and the associated cellular dynamics models.
Figure 2.
The nephron user interface showing the content tree on the left, graphical view in the centre and information panel on the right.
The content tree provides both an overview of the entire comprehensive model description and the primary model description navigation. The actual content of the tree is generated by defining a suitable query to perform against the comprehensive model description. In this manner, the presentation of the overview can be altered to suit specific purposes with no change required in either the model description or software tools. In the content tree example shown in figure 2, we present first the anatomy of the nephron by way of the various tubule segments and then the physiological function of each of these segments. These two sections allow the user to browse information about the underlying models, which includes information such as descriptions of specific anatomical structures, the cellular models associated with each segment and simulation results demonstrating the validity of specific transport protein models (matching published results, for example). The final section in the example content tree presents scientific investigations in which some (or all) of the models presented in the first two sections are instantiated in specific computational scenarios as part of the investigation.
In our prototype presentation, the user can navigate to and select an item of interest from the content tree. This item selection results in the interface displaying the information, which matches the selected item—which may be either graphical, rich text or a combination of both. Graphical information is displayed in the graphical view panel, such as a change in the highlighted segment, a new diagram being presented to the user, a specific spatially distributed field being visualized, or a change in viewing angle of a three-dimensional anatomical model. Rich text data are displayed in a new panel within the information panel.
As the name implies, the graphical view provides the user with a graphical presentation of relevant aspects of the comprehensive model description. In the current prototype configuration, we make use of two interactive graphical presentation technologies: a three-dimensional field visualization and manipulation tool for visualizations at the nephron spatial scale; and scalable vector graphics (SVG) diagrams for cellular and subcellular visualizations. The three-dimensional viewer provides the user with a graphical tool to navigate the model description via a stylized nephron diagram (as shown in figure 2) and also to visualize simulation results and experimental data (figure 3). This visualization tool allows the user to select items of interest, as does the content tree, in order to navigate to associated data from the model description.
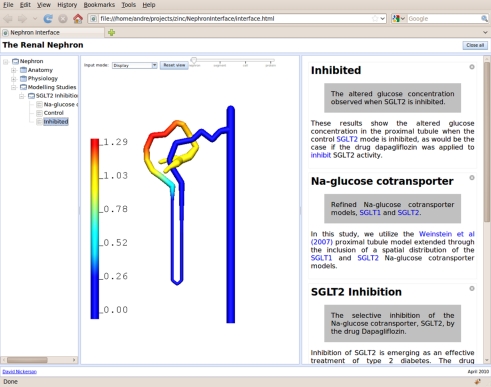
Figure 3.
An illustration of the user interface presenting a field-based visualization of simulation results from one of the modelling studies included in the comprehensive model description. In this example, the visualized field is showing the relative concentration of glucose in the lumen of the proximal nephron with the non-dimensional scale shown on the left in the graphical view panel (centre screen).
SVG is used to provide a graphical presentation tool for cellular and subcellular spatial scale information. In this, we are able to leverage the native support for SVG in most common web browsers to provide the user with a flexible and interactive visualization environment for the two-dimensional diagrams typically seen in descriptions of models at these spatial scales. As for the three-dimensional viewer discussed above, the user is able to navigate the model description by selecting items of interest in the various diagrams.
A ‘spatial scale’ slider (seen at the top of the graphical view in figure 2) is provided to allow the user to navigate between the spatial scales included in the comprehensive model description. When the user moves the slider, the user interface will move up or down the spatial scale hierarchy based on the currently selected item of interest. For example, selecting a particular transporter protein at the cell spatial scale and ‘zooming in’ will change the graphical view to show further data on that specific transporter protein. An example illustrating this is shown later in figure 4 with the transition from figure 4d to 4f. Alternatively, ‘zooming out’ would jump from the cellular view to displaying the corresponding nephron tubule in the three-dimensional viewer.
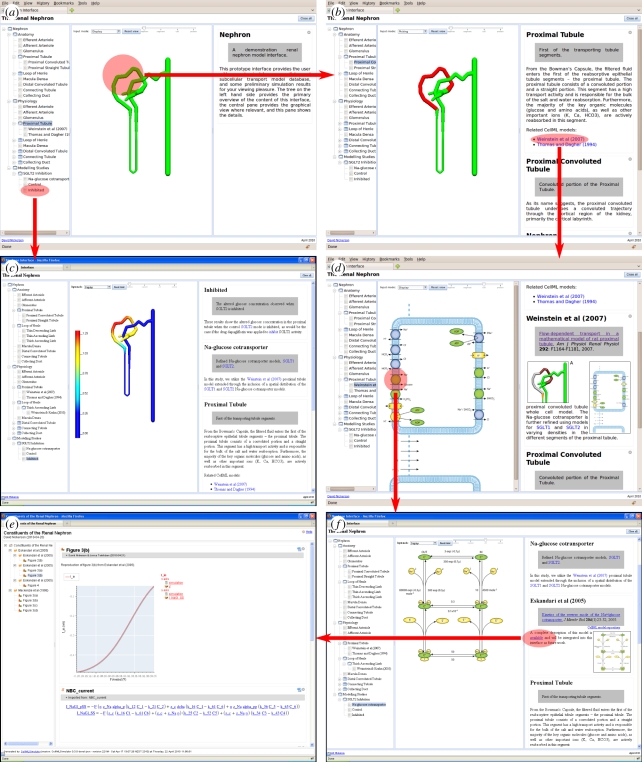
Figure 4.
Demonstration of a user navigating through an example modelling study using our prototype user interface. The highlighted regions indicate a user selection in one view resulting in a transition to the following view indicated by the arrows. See text for further details. The reader is invited to follow this sequence of steps themselves at http://www.abi.auckland.ac.nz/nephron/.
As demonstrated in figures 2 and 3, the information panel in our prototype interface provides the rendering of rich text data extracted from the comprehensive model description. Owing to developing the user interface for use in web browsers, we are again able to leverage the functionality common to all browsers and populate the rich text with hyperlinks to establish connections between related pieces of data. Such cross references can be both explicitly stated in the model description or implied by the software extracting the data from the underlying encoded models and associated annotations. Thus, we provide a third method for the user to navigate through the complete comprehensive model description. In combination, we believe that having these multiple methods by which the data can be navigated enhances the usability of the tool. This approach also establishes a framework that can be easily customized to emphasize specific routes through the data, a useful feature in some applications of this tool.
The interactive presentation of the nephron model we are developing allows the user to browse relevant aspects of nephron anatomy using the content tree, the stylized nephron diagram, or directly navigating between data items in the information panel. In practice, we find that a user will invariably use a combination of all these methods of navigation. As the user navigates the models exposed via the interface, they are directed to community model repositories (e.g. the CellML model repository, [14]), when appropriate, where they can obtain further information about the models. In addition to providing the user with a detailed and interactive description of the complete multi-scale nephron model, access to the results of simulation experiments is enabled by our tool. Currently available simulation experiments range from validation studies of transport mechanisms through to the effect of modifying transport dynamics in nephron segment models.
2.2. Comprehensive model description
Previous work has demonstrated the versatility of Physiome/VPH model encoding and annotation formats and methods in capturing the creation, evolution and application of models of cellular and subcellular physiology [8,14,15]. We have also discussed how such technologies can be extended to larger spatial scale models [13]. The current work provides an excellent ‘use case’ for helping further the development of the formats previously discussed (i.e. FieldML), associated annotation methods, and the framework for coupling and integrating the various formats into a cohesive comprehensive description of the model. Furthermore, the ever improving quality and scope of freely available scientific databases and ontologies (e.g. [14,16–19]) provides ample data for the detailed annotation of the mathematical models across all spatial scales.
In this work, we are using all these technologies to create a comprehensive description of a multi-scale renal nephron model. This comprehensive description includes both the definition of the mathematical models and the parameterization of them into specific simulation experiments. This comprehensive description includes models of individual transport proteins (e.g. [1,20,21]), whole cells (e.g. [22–24]) and nephron tubules (e.g. [22,25]). While we are still in the prototype and technology development phase, the level of detail encapsulated at each of the spatial scales varies, as does the completeness of the model annotations for the different constituent models. For example, some protein transport and cell models have been instantiated into simulation experiments representing the full range of published data for the models whereas others have only been minimally validated to ensure the model encoding represents the intended physiological function.
This work is at the leading edge of the model encoding and annotation formats developed under the Physiome Project/VPH umbrella. As such, we are helping guide the development of relevant formats, model annotation and best practice guidelines in the application of these technologies to the realm of computational physiology. The user interface we are developing in the current work provides an excellent consumer of these community efforts. We are thus able to provide detailed input and expertise to the development and maintenance of relevant software and infrastructural support.
2.3. Implementation
As mentioned above, our tool is a web application delivered to the user over the Internet using the Firefox web browser. The actual interface is based on the Dojo Toolkit (http://www.dojotoolkit.org), which provides the underlying technology for laying out the interface and linking the various interactive components together. The Zinc extension for Firefox (http://www.cmiss.org/cmgui/zinc) is used to provide the interactive three-dimensional nephron model viewer. While the Dojo Toolkit works across all major web browsers and operating systems, the version of Zinc used in this interface prototype is only available for Firefox. Work is currently underway at the Auckland Bioengineering Institute to extend support of the Zinc extension across a wider range of web browsers and operating systems.
The actual web interface we use in this work originates with the CellMLSimulator (http://cellml.sourceforge.net) tool that was specifically developed as a test application for the comprehensive model description technologies described previously [7,8]. Specifically, the Dojo presentation module of CellMLSimulator is integrated directly into the prototype model presentation interface. Furthermore, CellMLSimulator itself is used to generate the content for the parts of the renal nephron model description, which is encoded in CellML. This generated content is then extended with the hand-crafted portions for the sections of the nephron model that are not able to be expressed in CellML or associated annotations.
The Zinc Firefox extension provides a web-friendly interface to the cmgui software environment (http://www.cmiss.org/cmgui/). The three-dimensional stylized nephron presented to the user in our model description interface is currently encoded in the native cmgui file format. When the current prototype FieldML support in cmgui propagates through to the Zinc extension, the nephron anatomical model will be migrated to FieldML. The combination of CellML, FieldML and standardized annotations will greatly enhance the opportunities to share our multi-scale renal nephron model with the VPH community. In particular, many tools within the VPH Toolkit [26,27] will support these technologies.
3. Results
We have implemented a multi-scale computational model of the renal nephron segments based on previous models available from the literature at the level of individual transport proteins [1,20,21], whole cell [22,23] and nephron tubule [25]. In parallel to developing this computational model, we have defined a comprehensive model description of each of the constituent models and their assembly into various specific simulation experiments. This comprehensive model description is available at http://www.abi.auckland.ac.nz/nephron/. For the submodels encoded in CellML and associated annotation formats, we have followed the method described by Nickerson et al. [8] to generate the content for the interactive user interface. For the integrated tubule models, we have created custom software for performing simulation experiments with these models. This custom software is being used to inform the development of related formats and software tools under the Physiome/VPH umbrella and once these formats and tools are capable the models will be migrated to make use of them.
In a preliminary demonstration of both the nephron model implementation and the user interface, we have performed an initial simulation study investigating the role of the sodium–glucose cotransporters in the PT. Inhibition of the sodium–glucose cotransporter isoform SGLT2 is emerging as an effective treatment of type 2 diabetes [3,5,28]. The drug dapagliflozin binds competitively to SGLT2, inhibiting the reabsorption of glucose into the blood [5,28,29], and thus resulting in an increased excretion of glucose in the urine. Kinetic models of the sodium–glucose cotransporter isoforms (SGLT1 [1] and SGLT2 ([20], since shown to be pig SGLT3 [30] but used as a surrogate for SGLT2 in this initial demonstration)) were incorporated into the Weinstein et al. [23] model of the PT. Simulation of PT transport in the presence of dapagliflozin (inhibition of SGLT2) demonstrated an increased proportion of glucose remaining in the luminal solution to be ultimately excreted in the urine.
The mathematical models of nephron segments (and associated simulation results) are embedded in a one-dimensional finite-element model of the nephron. In figure 4, we demonstrate a user session in which a user navigates through this modelling study.
Figure 4a shows the initial screen that a user would see upon first loading the comprehensive nephron model description into their web browser. From this starting screen, and following the horizontal arrow, the user selects the PT from the three-dimensional nephron viewer. This action results in the information related to the PT segment model being displayed in the information panel, as shown in figure 4b. Part of the information displayed to the user for this segment is a listing of related CellML models that are part of the comprehensive nephron model description, which have been tagged as relevant to the PT segment. The user then chooses to select one of these models (in this example, the Weinstein et al. [23] model), which results in the user being presented with both further information about this cellular model in the information panel and a schematic diagram of all the components of this model in the graphical view (figure 4d). Upon examining the data now presented, the user decides to further investigate the sodium–glucose cotransporter. Thus, selecting the appropriate transport glyph in the schematic diagram, the user is now presented with the relevant information from the comprehensive description of the nephron model. As for the previous step, this user action has resulted in a change to the graphical view and new information being added to the information panel, shown in figure 4f. The graphical view now shows the state transition diagram for a specific sodium–glucose cotransporter model (SGLT1, [1]) that has been incorporated in the Weinstein et al. [23] cellular model.
From the description of the Eskandari et al. [1] model presented to the user in figure 4f, the user is able to discern that there is further information available for this particular model. The user then chooses to pursue this information and is presented with the full reference description for this model as shown in figure 4e (as per [8]). This description of the model allows the user to browse the mathematical model and simulation results related to this specific transport protein model. The Eskandari et al. [1] model is actually a model of SGLT1, which we have implemented as part of a study in to glucose transport in the PT. By moving back up the spatial scale, the user is able to observe the effect of this particular transport protein on glucose transport in the PT, as indicated by the vertical arrow in figure 4a and the resulting view of simulation results in figure 4c. Contrasting the results presented in figure 4c for the case where the SGLT2 surrogate is inhibited with the control simulation which is also available from figure 4a, the user is able to gain understanding into the role of SGLT2 in the maintenance of glucose homeostasis.
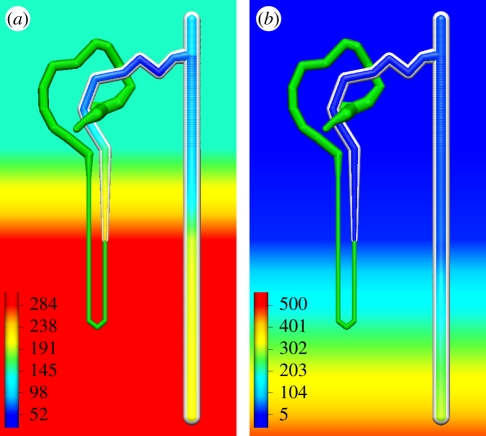
In a further demonstration of the capability of our comprehensive model description technology and the nephron model, which we are developing, figure 5 illustrates simulation results examining the behaviour of the thick ascending limb and distal portion of the nephron (reproducing the work of Weinstein [25]). In this model, spatial gradients of ion and solute concentrations in the bathing media surrounding the nephron (as demonstrated by the gradients of sodium and urea shown in the background of figure 5a,b, respectively) impact on the function of the epithelial cells, and thus the concentration gradients in the lumen of the nephron. These spatial gradients have been rendered so that the user is able to visualize the model results in the context of the boundary conditions and nephron model definition. We envision that in future versions of our tool, users would be able to interact with both the boundary conditions and model definition in order to investigate beyond the data captured in the comprehensive model description. For example, observing the change in luminal sodium concentration when changing the gradient in the bathing media or altering the distribution of transport proteins in certain tubule segments. Such functionality would greatly enhance the utility of this tool as a teaching aid.
Figure 5.
Results illustrating the use of a spatial gradient of solute concentrations in the bathing media interacting with the transport mechanisms in the nephron. (a) Sodium; (b) urea; scale bar as shown in each image in units of mmol l−1.
4. Conclusions
We have developed a framework for the comprehensive description of biophysically detailed multi-scale physiological models. Where possible, we use suitable community defined formats and technologies to represent the mathematical models and associated annotations. For the portions of the multi-scale model, which are not able to be represented using existing formats, we have developed custom methods for representing the data. These custom methods are now being used to help develop community standards within the Physiome/VPH projects in order to ensure our comprehensive model descriptions are entirely represented using community defined formats. This will greatly improve the ability to share and reuse models expressed using this framework among the scientific community.
In a demonstration of our model description framework, we have implemented a multi-scale computational model of the renal nephron segments. Using this model, we have been able to reproduce simulation experiments from the literature at the transport protein, whole cell and nephron tubule spatial scales. We have also performed some preliminary investigations using this model.
We have also constructed a prototype user interface that is able to present the comprehensive description of the multi-scale nephron model in an interactive web-based environment. We are currently developing both the nephron model and the user interface to include more functional segments of the nephron and the associated ion transport kinetics. Work is also underway to better integrate reference descriptions of the CellML models [7,8] within the overall user interface design, including the interactive pathway and cellular model diagrams. A tighter coupling with the CellML model repository, and potentially the geometric models soon to be available in the Physiome model repository (http://models.physiomeproject.org), is also highly desirable. Such coupling, however, will rely on greater access to the model repositories via clearly defined public interfaces and web services. Requirements such as this which arise during the development and application of our model description framework and software tools provide impetus for the development of the core Physiome Project/VPH software infrastructure.
With the development of greater software level access to the various model repositories and as the repository curators increase the level of annotation of the models therein, there is scope to enable our web-based presentation environment to directly access the models. This would greatly increase the flexibility for users of our web environment to interact with the multi-scale models. In future versions of our interface, users will be able to edit the model descriptions which form the comprehensive model description, changing boundary conditions, for example. Furthermore, with access to the model repositories, it would be possible to perform queries for alternative models that could be automatically substituted into the multi-scale model.
Acknowledgements
This project is funded by a Vice Chancellor's Strategic Development Fund from The University of Auckland. J.T. is supported by an Auckland Doctoral Scholarship. K.L.H. is supported by the Department of Physiology, University of Otago.
Footnotes
One contribution of 17 to a Theme Issue ‘The virtual physiological human’.
References
- 1.Eskandari S., Wright E. M., Loo D. D. 2005. Kinetics of the reverse mode of the Na+/glucose cotransporter. J. Membr. Biol. 204, 23–32 10.1007/s00232-005-0743-x (doi:10.1007/s00232-005-0743-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hummel C. S., Lu C., Loo D. D. F., Hirayama B. A., Voss A. A., Wright E. M. 2011. Glucose transport by human renal Na+/D-glucose cotransporters SGLT1 and SGLT2. Am. J. Physiol. Cell Physiol. 300, C14–C21 10.1152/ajpcell.00388.2010 (doi:10.1152/ajpcell.00388.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright E. M., Hirayama B. A., Loo D. F. 2007. Active sugar transport in health and disease. J. Intern. Med. 261, 32–43 10.1111/j.1365-2796.2006.01746.x (doi:10.1111/j.1365-2796.2006.01746.x) [DOI] [PubMed] [Google Scholar]
- 4.Parent L., Supplisson S., Loo D. D., Wright E. M. 1992. Electrogenic properties of the cloned Na+/glucose cotransporter. II. A transport model under nonrapid equilibrium conditions. J. Membr. Biol. 125, 63–79 10.1007/BF00235798 (doi:10.1007/BF00235798) [DOI] [PubMed] [Google Scholar]
- 5.Idris I., Donnelly R. 2009. Sodium–glucose co-transporter-2 inhibitors: an emerging new class of oral antidiabetic drug. Diabetes Obes. Metab. 11, 79–88 10.1111/j.1463-1326.2008.00982.x (doi:10.1111/j.1463-1326.2008.00982.x) [DOI] [PubMed] [Google Scholar]
- 6.Cuellar A. A., Lloyd C. M., Nielsen P. F., Bullivant D. P., Nickerson D. P., Hunter P. J. 2001. An overview of CellML 1.1, a biological model description language. Simulation 79, 740–747 10.1177/0037549703040939 (doi:10.1177/0037549703040939) [DOI] [Google Scholar]
- 7.Nickerson D., Buist M. 2008. Interactive reference descriptions of cellular electrophysiology models. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2008, 2427–2430 [DOI] [PubMed] [Google Scholar]
- 8.Nickerson D. P., Corrias A., Buist M. L. 2008. Reference descriptions of cellular electrophysiology models. Bioinformatics 24, 1112–1114 10.1093/bioinformatics/btn080 (doi:10.1093/bioinformatics/btn080) [DOI] [PubMed] [Google Scholar]
- 9.Cooling M. T., Rouilly V., Misirli G., Lawson J., Yu T., Hallinan J., Wipat A. 2010. Standard virtual biological parts: a repository of modular modeling components for synthetic biology. Bioinformatics 26, 925–931 10.1093/bioinformatics/btq063 (doi:10.1093/bioinformatics/btq063) [DOI] [PubMed] [Google Scholar]
- 10.Nickerson D., Buist M. 2008. Practical application of CellML 1.1: the integration of new mechanisms into a human ventricular myocyte model. Prog. Biophys. Mol. Biol. 98, 38–51 10.1016/j.pbiomolbio.2008.05.006 (doi:10.1016/j.pbiomolbio.2008.05.006) [DOI] [PubMed] [Google Scholar]
- 11.Schmid H., Nash M. P., Young A. A., Hunter P. J. 2006. Myocardial material parameter estimation-a comparative study for simple shear. J. Biomech. Eng. 128, 742–750 10.1115/1.2244576 (doi:10.1115/1.2244576) [DOI] [PubMed] [Google Scholar]
- 12.Christie G. R., Nielsen P. M., Blackett S. A., Bradley C. P., Hunter P. J. 2009. FieldML: concepts and implementation. Phil. Trans. R. Soc. A 367, 1869–1884 10.1098/rsta.2009.0025 (doi:10.1098/rsta.2009.0025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickerson D. P., Buist M. L. 2009. A physiome standards-based model publication paradigm. Phil. Trans. R. Soc. A 367, 1823–1844 10.1098/rsta.2008.0296 (doi:10.1098/rsta.2008.0296) [DOI] [PubMed] [Google Scholar]
- 14.Lloyd C. M., Lawson J. L., Hunter P. J., Nielsen P. F. 2008. The CellML model repository. Bioinformatics 24, 2122–2123 10.1093/bioinformatics/btn390 (doi:10.1093/bioinformatics/btn390) [DOI] [PubMed] [Google Scholar]
- 15.Beard D. A., et al. 2009. CellML metadata standards, associated tools and repositories. Phil. Trans. R. Soc. A 367, 1845–1867 10.1098/rsta.2008.0310 (doi:10.1098/rsta.2008.0310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M., et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 10.1038/75556 (doi:10.1038/75556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hines M. L., Morse T., Migliore M., Carnevale N. T., Shepherd G. M. 2004. ModelDB: a database to support computational neuroscience. J. Comput. Neurosci. 17, 7–11 10.1023/B:JCNS.0000023869.22017.2e (doi:10.1023/B:JCNS.0000023869.22017.2e) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laibe C., Le Novère N. 2007. MIRIAM Resources: tools to generate and resolve robust cross-references in Systems Biology. BMC Syst. Biol. 1, 58. 10.1186/1752-0509-1-58 (doi:10.1186/1752-0509-1-58) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C., et al. 2010. BioModels database: an enhanced, curated and annotated resource for published quantitative kinetic models. BMC Syst. Biol. 4, 92. 10.1186/1752-0509-4-92 (doi:10.1186/1752-0509-4-92) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackenzie B., Loo D. D., Panayotova-Heiermann M., Wright E. M. 1996. Biophysical characteristics of the pig kidney Na+/glucose cotransporter SGLT2 reveal a common mechanism for SGLT1 and SGLT2. J. Biol. Chem. 271, 32 678–32 683 10.1074/jbc.271.8.4086 (doi:10.1074/jbc.271.8.4086) [DOI] [PubMed] [Google Scholar]
- 21.Weinstein A. M. 2010. A mathematical model of rat ascending Henle limb. III. Cotransporter function. Am. J. Physiol. Renal. Physiol. 298, F512–F524 10.1152/ajprenal.00230.2009 (doi:10.1152/ajprenal.00230.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang H., Fujita T. 1999. A numerical model of the renal distal tubule. Am. J. Physiol. 276, F931–F951 [DOI] [PubMed] [Google Scholar]
- 23.Weinstein A. M., Weinbaum S., Duan Y., Du Z., Yan Q., Wang T. 2007. Flow-dependent transport in a mathematical model of rat proximal tubule. Am. J. Physiol. Renal. Physiol. 292, F1164–F1181 10.1152/ajprenal.00392.2006 (doi:10.1152/ajprenal.00392.2006) [DOI] [PubMed] [Google Scholar]
- 24.Weinstein A. M., Krahn T. A. 2010. A mathematical model of rat ascending Henle limb. II. Epithelial function. Am. J. Physiol. Renal. Physiol. 298, F525–F542 10.1152/ajprenal.00231.2009 (doi:10.1152/ajprenal.00231.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein A. M. 2010. A mathematical model of rat ascending Henle limb. III. Tubular function. Am. J. Physiol. Renal. Physiol. 298, F543–F556 10.1152/ajprenal.00232.2009 (doi:10.1152/ajprenal.00232.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper J., et al. 2010. The Virtual Physiological Human ToolKit. Phil. Trans. R. Soc. A 368, 3925–3936 10.1098/rsta.2010.0144 (doi:10.1098/rsta.2010.0144) [DOI] [PubMed] [Google Scholar]
- 27.Garny A., Cooper J., Hunter P. J. 2010. Toward a VPH/Physiome ToolKit. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 134–147 10.1002/wsbm.63 (doi:10.1002/wsbm.63) [DOI] [PubMed] [Google Scholar]
- 28.Kipnes M. 2009. Dapagliflozin: an emerging treatment option in type 2 diabetes. Expert Opin. Invest. Drugs 18, 327–334 10.1517/13543780902766794 (doi:10.1517/13543780902766794) [DOI] [PubMed] [Google Scholar]
- 29.Meng W., et al. 2008. Discovery of dapagliflozin: a potent, selective renal sodium-dependent glucose cotransporter 2 (SGLT2) inhibitor for the treatment of type 2 diabetes. J. Med. Chem. 51, 1145–1149 10.1021/jm701272q (doi:10.1021/jm701272q) [DOI] [PubMed] [Google Scholar]
- 30.Wright E. M. 2001. Renal Na+-glucose cotransporters. Am. J. Physiol. Renal Physiol. 280, F10–F18 [DOI] [PubMed] [Google Scholar]