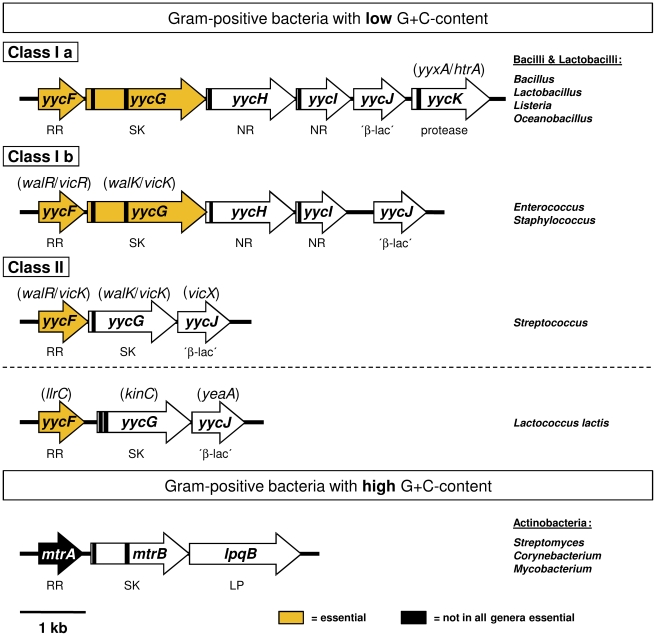
Figure 1. Gram-positive bacteria with orthologous yyc (wal/vic) genes.
The upper part of the figure shows the structural organization of orthologous yyc operons in Gram-positive bacteria with low G+C-content adopting the classification into class I and II proposed by Szurmant et al. [5]. To distinguish between genera that contain 5 or 6 cistrons, the class I operon was divided into two subclasses. For a clear overview, orthologous genes are named according to the corresponding yyc gene in Bacillus subtilis in all genera and synonymous terms commonly used in a genus are given in brackets. The lower part of the figure illustrates the organization of the mtrAB/lpqB operon in Gram-positive Actinobacteria with high G+C-content. The operons are drawn to scale from representative species: Bacillus subtilis 168 (NC_000962), Staphylococcus aureus N315 (NC_002745), Streptococcus pneumoniae R6 (NC_003098), Lactococcus lactis Il1403 (NC_002662), Corynebacterium glutamicum ATCC 13032 (NC_006958). Essential genes are highlighted. TM coding regions were determined utilizing the TMHMM 2.0 server 2.0 web interface (http://www.cbs.dtu.dk/services/TMHMM/) and marked as black bars. Functions are indicated below the arrows: RR = response regulator, SK = sensor kinase, NR = negative regulator of kinase function, ‘β-lac’ = similarities to an enzyme super-family containing metallo-β-lactamases, protease = serine protease, LP = conserved lipoprotein. Within the class II of yyc operons, the YycG kinase of Lactococcus lactis is an exception of the rule, because it possesses two transmembrane domains (instead of one), which flank a very short extracytoplasmic loop comprising four amino acids [7], [8].