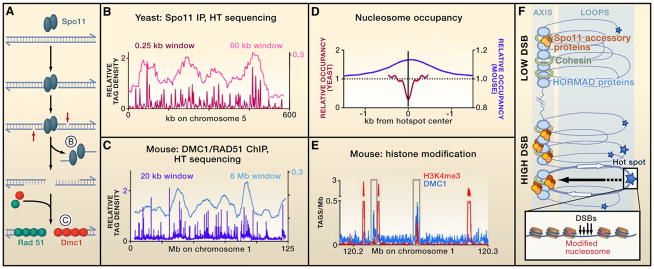
Figure 1. Genome-wide Features of Meiotic Double-Strand Breaks.
(A) Mechanisms of meiotic DNA double-strand break (DSBs) formation, showing the substrates that Pan et al. (2011) and Smagulova et al. (2011) used to map DSBs in yeast and mouse, respectively.
(B and C) DSB distributions on yeast and mouse chromosomes, with smoothing windows to show individual DSB hot spots and DSB domains.
(D) Nucleosome occupancy differs at yeast and mouse hot spots.
(E) Mouse hot spots are enriched for the trimethylation on lysine 4 of histone H3 (H3K4me3).
(B–E) These panels are plotted using data of Pan et al. (2011) and Smagulova et al. (2011). In (B) and (C), tag densities are normalized to relative genome size.
(F) Chromatin and chromosome organization influence DSB formation. DSBs occur within loops, within or outside genes (white arrows), in accessible chromatin where nucleosomes carry specific modifications and at sites where Spo11 is recruited (stars indicate different frequencies of DSBs). Some Spo11 accessory proteins (brown/orange ovals) are localized in axes, suggesting that DSB formation can involve the movement of loop sites to the axis.