Abstract
Marine sponges have been recognized as potentially rich sources of various bioactive molecules. In our continuing search for new secondary metabolites from Indonesian marine invertebrates, we collected a sponge, whose extract showed cytotoxicity against cultured cells at 0.1 μg/mL. Purification of the extract yielded two new macrolides 2 and 3 along with known candidaspongiolide (1). The structures for compounds 2 and 3 were elucidated by spectral analysis (1H, 13C, COSY, HMQC, HMBC) and by comparison of their NMR data with those of 1. Compounds 2 and 3 exhibited a little more potent cytotoxicity (IC50 4.7 and 19 ng/mL) than that (IC50 37 ng/mL) of candidaspongiolide (1) against NBT-T2 cells.
1. Introduction
Sponges, a group of sedentary organisms, cannot move and escape from predators. Most sponges are filter feeders pumping water to its body to obtain foods and oxygen and to expel wastes and may be threatened by microorganisms during filtering seawater rich in bacteria and fungi [1, 2]. In order to defend themselves against predators, pathogens and competitors, sponges may have developed to produce or accumulate secondary metabolites during their long evolution, such as feeding deterrent, antimicrobial, antifungal, and antifouling molecules. Interestingly, some of the compounds have also shown remarkable potency as drug candidates against various human diseases as discussed elsewhere [3–9].
In 1984, Schmitz and coworkers isolated tedanolide from the Caribbean marine sponge Tedania ignis [10]. Tedanolide is a unique 18-membered macrolide where lactonization occurs at a primary hydroxyl group instead of a common secondary one, and this class of macrolide has been reported to exhibit strong cytotoxicity at pico to nanomolar range [10, 11]. The unique structure in combination with promising biological activity leads tedanolide as an intriguing target for formal and total syntheses [12–14]. More recently, Meragelman and coworkers reported a macrolide named candidaspongiolide (1) related to tedanolide with modification at C-11 to C-15 from the marine sponge Candidaspongia sp. Candidaspongiolide exhibited potent cytotoxicity in NCI 60 cells panel with GI50 of 14 ng/mL [15], protein synthesis inhibition, and apoptosis induction [16].
In our continuing search for potential drug leads from Indonesian marine invertebrates [17, 18], we obtained a sponge whose extract showed cytotoxicity at 0.1 μg/mL against NBT-T2 cells in a screening process. Purification of the extract provided candidaspongiolide (1) along with two new analogs 2 and 3, which are the subject of this paper.
2. Materials and Methods
2.1. Chemicals and Equipments
Methanol (MeOH) used for extraction was of technical grade. Reagent grade solvents were used for isolating compounds 1–3. Merck Si-60 (70–230 mesh) was used for silica gel column chromatography, while Merck Si-60 F254 for analytical TLC. HPLC was performed either on a Waters 510 pump with a Waters 486 UV detector and a Shodex RI-101 or on a Hitachi L-6000 pump with a Hitachi L-4000 UV detector and a Shodex RI-101 using a Mightysil Si-60 (10 × 250 mm) column. Optical rotations were measured on a Jasco P-1010 polarimeter using a cell with 3.5 mm aperture. IR spectra were recorded on a Jasco FT/IR-6100 instrument, whereas HRESIMS was measured on a Jeol JMS-T100LP spectrometer using reserpine or sodium trifluoroacetate as an internal standard. Most of 1H and 13C NMR spectra were measured in CDCl3, while those of compound 3 were measured in CD3OD with TMS as an internal standard on a Jeol A500 and/or a Bruker AVANCE III-500 in CDCl3. The 1H and 13C chemical shifts were given in ppm, while coupling constants were in Hz.
2.2. Sponge
Specimens of the sponge tagged K09-02 was collected by hand using SCUBA at 15–25 m depth at Kupang, West Timor, East Nusa Tenggara, Indonesia on August 2009. By comparing underwater images of our specimen with that of the specimen of NCI group [15], it is likely to be the same sponge. The specimen was kept frozen until extraction. The sponge K09-02 may be an endemic species to this region. The colonies are grey in color and stand.
2.3. Extraction and Isolation
After cutting into small pieces, the sponge (653 g, wet) was soaked in MeOH for 24 h for three times. Then, the solution was concentrated under vacuum to obtain a crude extract. The methanolic extract (17.0 g) was triturated with ethyl acetate (EtOAc) to provide a lipophilic fraction (2.7 g), which killed NBT-T2 cells at 0.1 μg/mL. This fraction was subjected to a silica gel column eluting with stepwise gradient solvents (hexane : EtOAc = 2 : 1, 1 : 1, 1 : 2, 0 : 1, EtOAc : MeOH = 10 : 1) to afford ten fractions. Fraction 5 (126.0 mg) was purified by repetitive Si-60 HPLC using hexane-EtOAc mixtures to provide candidaspongiolide 1 (15.4 mg). Fraction 6 (70.8 mg) was also purified by Si-60 HPLC using a solvent system hexane : EtOAc = 2 : 1 to afford compound 2 (9.8 mg). Fraction 9 (107.1 mg) was also subjected to repetitive Si-60 HPLC using hexane : EtOAc = 1 : 6, EtOAc : CH2Cl2 : MeOH = 20 : 20 : 1, and EtOAc : CH2Cl2 : MeOH = 10 : 20 : 1 as solvent systems sequentially, to give compound 3 (21.8 mg). Isolation of these compounds was guided by cytotoxicity testing and NMR spectra.
2.4. Compound 1
Colorless glass, [α]D 25 +69 (c 0.55, MeOH). IR ν max (neat) 3419, 2925, 2854, 1742, 1715, 1456, 1372, 1234, 1086 cm−1. 1H and 13C NMR; see Tables 1 and 2. HR-ESIMS [M+Na]+ m/z 945.55514, 959.57079, 973.58884 (calcd for C50H82NaO15 + 945.55459 (Δ +0.58 ppm), C51H84NaO15 + 959.57024 (+0.57 ppm), and C52H86NaO15 + 973.58589 (+3.0 ppm)).
Table 1.
13C NMR data for compounds 1, 2, and 3.
| C no. | 115,a | 1a | 2a | 3b |
|---|---|---|---|---|
| 1 | 171.3 qC | 171.3 qC | 171.3 qC | 171.3 qC |
| 2 | 70.7 CH | 70.8 CH | 70.8 CH | 72.9 CH |
| 3 | 83.4 CH | 83.3 CH | 83.1 CH | 84.7 CH |
| 4 | 47.8 CH | 47.7 CH | 47.9 CH | 49.5 CH |
| 5 | 214.6 qC | 214.9 qC | 216.2 qC | 217.6 qC |
| 6 | 48.5 CH | 48.3 CH | 49.8 CH | 51.1 CH |
| 7 | 80.1 CH | 80.1 CH | 79.2 CH | 79.9 CH |
| 8 | 131.6 qC | 132.0 qC | 136.2 qC | 139.0 qC |
| 9 | 132.0 CH | 131.7 CH | 129.4 CH | 129.8 CH |
| 10 | 46.2 CH | 46.1 CH | 45.9 CH | 46.5 CH |
| 11 | 211.5 qC | 211.7 qC | 211.9 qC | 212.7 qC |
| 12 | 42.6 CH2 | 42.7 CH2 | 42.5 CH2 | 44.3 CH2 |
| 13 | 68.9 CH | 68.8 CH | 69.0 CH | 69.3 CH |
| 14 | 81.6 qC | 81.6 qC | 81.6 qC | 85.0 qC |
| 15 | 210.8 qC | 211.0 qC | 211.2 qC | 216.5 qC |
| 16 | 46.9 CH | 47.0 CH | 46.9 CH | 46.4 CH |
| 17 | 77.8 CH | 77.7 CH | 77.7 CH | 78.4 CH |
| 18 | 62.7 qC | 62.7 qC | 62.6 qC | 63.9 qC |
| 19 | 67.1 CH | 67.0 CH | 67.0 CH | 67.4CH |
| 20 | 31.1 CH | 31.1 CH | 30.9 CH | 32.4 CH |
| 21 | 129.7 CH | 129.4 CH | 129.6 CH | 131.6 CH |
| 22 | 125.5 CH | 125.5 CH | 125.4 CH | 126.2 CH |
| 23 | 13.5 CH3 | 13.4 CH3 | 13.3 CH3 | 13.3 CH3 |
| 24 | 14.6 CH3 | 14.5 CH3 | 14.4 CH3 | 15.3 CH3 |
| 25 | 14.7 CH3 | 14.5 CH3 | 15.0 CH3 | 15.6 CH3 |
| 26 | 10.8 CH3 | 10.7 CH3 | 10.2 CH3 | 10.5 CH3 |
| 27 | 16.3 CH3 | 16.2 CH3 | 16.4 CH3 | 15.7 CH3 |
| 28 | 63.5 CH2 | 63.5 CH2 | 63.5 CH2 | 65.7 CH2 |
| 29 | 63.2 CH2 | 63.2 CH2 | 62.8 CH2 | 64.8 CH2 |
| 30 | 11.1 CH3 | 11.0 CH3 | 10.9 CH3 | 11.5 CH3 |
| 31 | 18.6 CH3 | 18.4 CH3 | 18.3 CH3 | 18.7 CH3 |
| 32 | 60.3 CH3 | 60.3 CH3 | 60.3 CH3 | 60.3 CH3 |
| 33 | 169.5 qC | 169.9 qC | — | — |
| 34 | 21.6 CH3 | 21.5 CH3 | — | — |
| 35 | 173.5 qC | 173.7 qC | 173.6 qC | — |
| 36 | 34.2 CH2 | 34.1 CH2 | 34.0 CH2 | — |
| 37 | 29.0 CH2 | 29.0 CH2 | 29.1 CH2 | — |
aMeasured in CDCl3. bMeasured in CD3OD.
Table 2.
1H NMR data for compounds 1, 2, and 3 (J in Hz).
| C no. | 115,a | 1a | 2a | 3b |
|---|---|---|---|---|
| 1 | — | — | — | — |
| 2 | 3.96 dd (1.0, 7.3) | 3.96 dd (1.3, 7.5) | 3.98 dd (1.3, 7.8) | 3.76 d (2.2) |
| 3 | 3.64 dd (1.3, 7.8) | 3.67 dd (1.3, 8.0) | 3.67 dd (1.3, 8.4) | 3.81 dd (2.2, 9.8) |
| 4 | 3.12 m | 3.13 dd (8.0, 7.1) | 3.10 dq (8.4, 7.3) | 3.16 dq (9.8, 7.1) |
| 5 | — | — | — | — |
| 6 | 3.18 dq (10.7, 7.3) | 3.22 dq (10.7, 6.8) | 3.04 dq (9.8, 6.8) | 3.16 dq (10.0, 7.1) |
| 7 | 5.39 d (10.7) | 5.41 d (10.7) | 4.12 d (10.0) | 4.03 d (10.0) |
| 8 | — | — | — | — |
| 9 | 5.60 d (9.3) | 5.62 d (9.6) | 5.48 d (10.5) | 5.33 d (9.3) |
| 10 | 3.38 dq (9.3, 6.8) | 3.41 dq (9.6, 7.0) | 3.49 dq (10.5, 7.1) | 3.36 dq (9.3, 6.8) |
| 11 | — | — | — | — |
| 12 | 2.66 dd (9.8, 16.1) | 2.69 dd (9.8, 16.1) | 2.72 dd (9.8, 16.1) | 2.75 dd (9.5, 17.6) |
| 2.49 dd (2.4, 16.1) | 2.49 dd (2.5, 16.1) | 2.51 dd (2.0, 16.1) | 2.23 dd (2.0, 17.6) | |
| 13 | 4.40 m | 4.42 m | 4.39 dt (2.0, 9.8) | 4.44 dd (2.9, 9.5) |
| 14 | — | — | — | — |
| 15 | — | — | — | — |
| 16 | 4.02 dt (3.9, 10.9) | 4.03 ddd (3.9, 10.5, 11.5) | 4.09 dt (3.9, 11.0) | 4.08 ddd (3.9, 10.8, 11.4) |
| 17 | 3.12 m | 3.13 m | 3.20 dd (11.0) | 3.20 d (10.7) |
| 18 | — | — | — | — |
| 19 | 2.56 d (9.3) | 2.59 d (9.3) | 2.58 d (9.8) | 2.62 d (9.3) |
| 20 | 2.44 m | 2.47 m | 2.47 m | 2.48 m |
| 21 | 5.23 dt (1.5, 10.7) | 5.25 ddd (0.7, 10.2, 10.9) | 5.24 dt (1.5, 10.5) | 5.31 m |
| 22 | 5.48 dq (10.7, 6.8) | 5.51 dq (10.9, 6.8) | 5.49 dq (10.5, 6.8) | 5.51 dq (10.7, 6.8) |
| 23 | 1.59 dd (1.5, 6.8) | 1.62 dd (1.2, 6.8) | 1.62 dd (1.5, 6.8) | 1.62 dd (1.7, 6.8) |
| 24 | 1.18 d (6.8) | 1.21 d (7.1) | 1.21 d (7.3) | 1.23 d (7.1) |
| 25 | 1.13 d (7.3) | 1.16 d (6.8) | 1.28 d (6.8) | 1.26 d (7.1) |
| 26 | 1.54 brd (1.0) | 1.59 d (0.8) | 1.63 s | 1.65 d (1.5) |
| 27 | 1.07 d (6.8) | 1.09 d (7.0) | 1.10 d (7.1) | 1.03 d (6.8) |
| 28 | 4.44 d (11.7) | 4.46 d (11.5) | 4.45 d (11.5) | 3.75 d (10.5) |
| 4.19 d (11.7) | 4.22 d (11.5) | 4.21 d (11.5) | 3.76 d (10.5) | |
| 29 | 4.17 dd (3.7, 9.8) | 4.20 dd (3.7, 10.2) | 4.24 dd (3.0, 9.5) | 4.35 dd (3.9, 10.5) |
| 4.10 dd (10.2, 10.9) | 4.12 dd (10.2, 11.2) | 4.08 m | 3.91 dd (10.5, 11.4) | |
| 30 | 1.38 s | 1.42 s | 1.42 s | 1.35 s |
| 31 | 1.11 d (6.4) | 1.13 d (6.3) | 1.14 d (6.6) | 1.11 d (6.6) |
| 32 | 3.28 s | 3.31 s | 3.30 s | 3.39 s |
| 33 | — | — | — | — |
| 34 | 2.01 s | 2.04 s | — | — |
| 35 | — | — | — | — |
| 36 | 2.24 t (7.6) | 2.27 t (7.6) | 2.27 t (11.8) | |
| 37 | 1.53 brs | 1.59 brs | 1.25 brs | |
| OH-2 | 2.85 d (7.3) | 2.92 d (7.5) | 2.99 d (7.8) | 2.92 d (7.5) |
| OH-13 | — | 4.71 s |
aMeasured in CDCl3. bMeasured in CD3OD.
2.5. Compound 2
Colorless glass, [α]D 25 +72 (c 0.75, MeOH). IR ν max (neat) 3421, 2925, 2854, 1748, 1715, 1456 cm−1. 1H and 13C NMR; see Tables 1 and 2. HR-ESIMS [M+Na]+ m/z 903.54964, 917.56023, 931.57576, 945.60036 and 959.61056 (calcd for C48H80NaO14 + 903.54403 (Δ +6.2 ppm), C49H82NaO14 + 917.55968 (+0.60 ppm), C50H84NaO14 + 931.57533 (+0.46 ppm), C51H86NaO14 + 945.59098 (+9.9 ppm), and C52H88NaO14 + 959.60663 (+4.1 ppm)).
2.6. Compound 3
Yellow glass, [α]D 25 +97 (c 0.35, MeOH). IR ν max (neat) 3418, 2925, 2854, 1715, 1457, 1373, 1244, 1084, 995 cm−1. 1H and 13C NMR; see Tables 1 and 2. HR-ESIMS [M+Na]+ m/z 665.31522 (calcd for C32H50NaO13 + 665.31436 (+1.3 ppm)).
2.7. Acetylation
Compound 1 (0.2 mg) was dissolved in pyridine (50 μL) and acetic anhydride (50 μL). The mixture was stirred for three days under a nitrogen atmosphere at room temperature. After removal of excess reagents with nitrogen flow and vacuum, the reaction product 4 was checked with 1H NMR and ESIMS. Compound 2 was similarly treated to give 4.
2.8. Compound 4 from 1
1H NMR: δ 5.65 dd (J = 2.7, 9.5 Hz), 5.52 m, 5.51 d (J = 10.1 Hz), 5.36 m, 5.34 dd (J = 2.2, 9.1 Hz), 5.28 dt (J = 2.3, 10.5 Hz), 5.07 d (J = 11.0 Hz), 4.79 d (J = 6.6 Hz), 4.54 dd (J = 2.0, 11.3 Hz), 4.25 dd (J = 2.3, 8.1 Hz), 4.13 dd (J = 6.3, 11.5 Hz), 3.43 s (3H), 3.34 dq (J = 10.0, 6.7 Hz), 3.29 dq (J = 10.5, 7.0 Hz), 3.01 dd (J = 9.2, 18.4 Hz), 2.88 d (J = 9.3 Hz), 2.60 dd (J = 2.6, 18.4 Hz), 2.35–2.4 m, 2.32 dd (J = 1.9, 9.3 Hz), 2.21 s (3H), 2.10 s (3H), 2.09 s (3H), 2.02 s (3H), 1.69 d (J = 1.1 Hz, 3H), 1.66 dd (J = 1.7, 6.8 Hz, 3H), 1.44 d (J = 7.1 Hz, 3H), 1.37 s (3H), 1.17 d (J = 7.1 Hz, 3H), 1.08 d (J = 6.6 Hz, 3H), 0.90 t (J = 6.1 Hz, 3H). HR-ESIMS m/z 1072.60225, 1086.60997, 1099.62465 (calcd for 12C55 13CH88NaO18 + 1072.59019 (+11.24 ppm), 12C56 13CH90NaO18 + 1086.60584 (+3.8 ppm), and C58H92NaO18 + 1099.61814 (+5.9 ppm)).
2.9. Compound 4 from 2
1H NMR: δ 5.65 dd (J = 2.6, 9.8 Hz), 5.52 m, 5.51 d (J = 10.2 Hz), 5.37 m, 5.34 dd (J = 1.7, 9.1 Hz), 5.28 dt (J = 1.7, 10.5 Hz), 5.07 d (J = 10.9 Hz), 4.78 d (J = 6.3 Hz), 4.54 dd (J = 3.1, 11.7 Hz), 4.25 dd (J = 2.3, 8.1 Hz), 4.13 dd (J = 6.5, 11.3 Hz), 3.43 s (3H), 3.34 dq (J = 10.1, 5.7 Hz), 3.29 dq (J = 10.5, 7.2 Hz), 3.01 dd (J = 9.4, 18.6 Hz), 2.88 d (J = 9.2 Hz), 2.60 dd (J = 2.5, 18.6 Hz), 2.35–2.4 m, 2.32 (J = 1.9, 9.3 Hz), 2.22 s (3H), 2.10 s (3H), 2.09 s (3H), 2.02 s (3H), 1.70 d (J = 1.1 Hz, 3H), 1.62 dd (J = 1.7, 6.8 Hz, 3H), 1.44 d (J = 7.1 Hz, 3H), 1.37 s (3H), 1.17 d (J = 7.1 Hz, 3H), 1.08 d (J = 6.6 Hz, 3H), 0.90 t (J = 6.1 Hz, 3H). HR-ESIMS [M+Na+] m/z 1071.58258, 1085.60199, 1099.61165 (calcd for C56H88NaO18 + 1071.58629 (−4 ppm), C57H90NaO18 + 1085.60194 (−.5 ppm), and C58H92NaO18 + 1099.61759 (−5.9 ppm)).
2.10. Screening Process
NBT-T2 cells were purchased from Riken and used for cytotoxicity testing. NBT-T2 is a cell line derived from chemically induced rat bladder carcinoma cells [19]. The sponge extract was tested at 0.1, 1, and 10 μg/mL in triplicate, while fractions were done at 0.01, 0.1, and 1 μg/mL. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with Sigma antibiotic-antimycotic, Biowest fetal bovine serum, Gibco MEM nonessential amino acid in a Falcon 24-well plate or 48-well plate. After adding the extract or a fraction, cells were incubated for 24 h under 5% CO2 at 36°C [16]. Then, the cells were observed under a microscope to evaluate viability of cells whether the fractions were cytotoxic or not.
2.11. MTT Assay
Cultured cells were inoculated to a 96-well plate with approximate cell density of 1 × 104 cells/mL in DMEM. After 24 h incubation, a series of DMSO solution of compounds 1–3 were applied to each well and the final concentrations were adjusted as 0, 1, 12.5, 25, 37.5, 50, 62.5, to 75 ng/mL. Cells were incubated for another 24 h, and the media were replaced with 20 μL of 5 g/mL MTT solution in PBS and incubated for 3.5 h. After removal of PBS solution, an amount of 150 μL of DMSO was added to each well and the cells were reincubated for 15 min prior to measurement with a Tecan microplate reader at 590 nm with reference filter at 620 nm [20, 21].
3. Results and Discussion
As an EtOAc soluble portion of a methanolic extract of the sponge K09-02 showed potent cytotoxicity against cultured NBT-T2 cells, the portion was separated repetitively on a silica gel column followed by Si-60 HPLC affording three compounds 1, 2, and 3 as shown in Figure 1.
Figure 1.
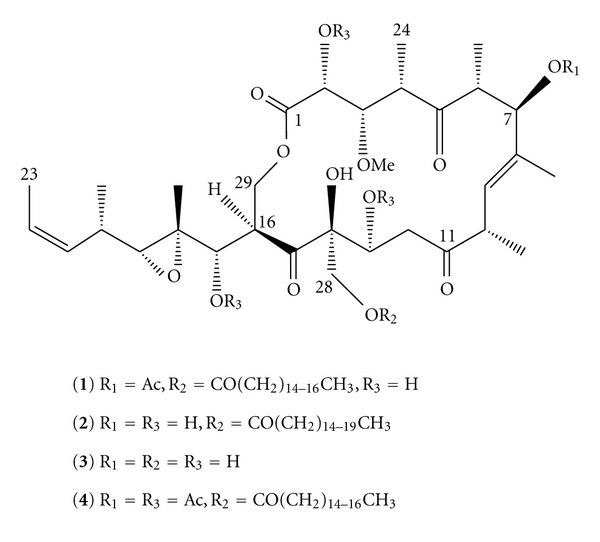
Structures of compounds (1)–(4).
By inspecting 1H and 13C NMR spectra of compound 1 together with database search (Tables 1 and 2, Figures S1 and S2 (Supplementary Materials available online at doi:10.5402/2011/852619)), we could readily identify that it is a member of candidaspongiolide, a series of 18-membered cytotoxic macrolide retaining one of fatty acid moieties from C14 to C18 at C-28 [15]. HR-ESIMS of our material exhibited molecular-related ions at m/z 945.55514, 959.57079, 973.58884 [M+Na]+ indicating that compound 1 is candidaspongiolide esterified with the homologs of three saturated fatty acids (palmitic, margaric, and stearic acids).
Compound 2 was obtained as a colorless glass with [α]D 25 +72. After elucidation of its 1H and 13C NMR spectra, compound 2 was found to be an analog of 1. However, the 13C NMR spectrum showed two carbonyl carbons at δ C 171.3 q (C-1) and 173.1 (C-35) instead of three in 1 (Table 1, Figure S3). As the signals for an acetoxy group (δ H 2.04 s, δ C 21.5 q) in 1 are missing in 2, it was suggested that 2 is a deacetyl derivative of 1. The lack of the acetyl group is in a good agreement with 1H NMR spectrum and COSY analysis showing that H-7 proton signal (δ H 4.12 d, J = 10.0 Hz) in 2 shifted to higher field than that (δ H 5.41 d, J = 10.7 Hz) in 1 (Table 2). HR-ESIMS of 2 showed a series of sodiated ions [M+Na]+ at m/z 903.54964, 917.56023, 931.57576, 945.60036, and 959.61056 corresponding to the presence of C16 to C20 esters. For structural confirmation, compound 2 was acetylated to give tetraacetate 4, which showed signals identical with 4 obtained from 1 (Figure S4). Compound 4 exhibited four acetyl signals at δ H 2.22 s, 2.10 s, 2.09 s, and 2.02 s and molecular-related ions corresponding to macrolide esters with C16 to C18 fatty acids.
Compound 3 was isolated as a yellowish glass with [α]D 25 +97. Its molecular formula was established as C32H50O13 by observing a molecular-related ion at m/z 665.31522 [M+Na]+ in HR-ESIMS. 1H and 13C NMR spectra (Tables 1 and 2, Figures S5 and S6) revealed that compound 3 has a similar macrolide structure to that of compound 1 except for the lack of a fatty acid ester moiety and an acetate found in 1. Higher field chemical shifts observed for H-7 (δ H 4.03) and H-28 (δ H 3.75) indicated that 3 is devoid of acyl groups. Close similarity of 1H and 13C NMR data of 3 to 1 (Table 2) indicated that the macrolide core structure of compound 3 is identical to compound 1.
All of natural compounds 1–3 exhibited potent cytotoxicity, IC50 37, 4.7, and 19 ng/mL, against NBT-T2 cells. The result is not in good agreement with those reported by Meragelman and coworkers, that is, candidaspongiolide (1) showed stronger growth inhibition (GI50 14 ng/mL) than the core compound (42 ng/mL) [15]. Additionally Paul et al. paperd the importance of a linear carbon chain on the cytotoxicity in the case of amphidinol [22]. The difference may be explained either by the number of cell lines or by different sensitivity of NBT-T2 cells.
Acknowledgments
The authors appreciate Professor Rob van Soest, University of Amsterdam, for showing a sponge image and suggestions on the sponge identification and Mr. T. Iha for mass measurements. they thank PharmaMar S. A. for financial support.
References
- 1.Ruzicka R, Gleason DF. Sponge community structure and anti-predator defenses on temperate reefs of the South Atlantic Bight. Journal of Experimental Marine Biology and Ecology. 2009;380(1-2):36–46. [Google Scholar]
- 2.Pfannkuchen M, Fritz GB, Schlesinger S, Bayer K, Brümmer F. In situ pumping activity of the sponge Aplysina aerophoba, Nardo 1886. Journal of Experimental Marine Biology and Ecology. 2009;369(1):65–71. [Google Scholar]
- 3.Assmann M, van Soest RWM, Köck M. New antifeedant bromopyrrole alkaloid from the caribbean sponge Stylissa caribica . Journal of Natural Products. 2001;64(10):1345–1347. doi: 10.1021/np000482s. [DOI] [PubMed] [Google Scholar]
- 4.Vik A, Hedner E, Charnock C, et al. (+)-Agelasine D: improved synthesis and evaluation of antibacterial and cytotoxic activities. Journal of Natural Products. 2006;69(3):381–386. doi: 10.1021/np050424c. [DOI] [PubMed] [Google Scholar]
- 5.Boonlarppradab C, Faulkner DJ. Eurysterols A and B, cytotoxic and antifungal steroidal sulfates from a marine sponge of the genus Euryspongia . Journal of Natural Products. 2007;70(5):846–848. doi: 10.1021/np060472c. [DOI] [PubMed] [Google Scholar]
- 6.Sjögren M, Göransson U, Johnson AL, et al. Antifouling activity of brominated cyclopeptides from the marine sponge Geodia barretti . Journal of Natural Products. 2004;67(3):368–372. doi: 10.1021/np0302403. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura S, Matsunaga S, Yoshida S, Nakao Y, Hirota H, Fusetani N. Structure-activity relationship study on 13-deoxytedanolide, a highly antitumor macrolide from the marine sponge Mycale adhaerens . Bioorganic and Medicinal Chemistry. 2005;13(2):455–462. doi: 10.1016/j.bmc.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Longeon A, Copp BR, Roué M, et al. New bioactive halenaquinone derivatives from South Pacific marine sponges of the genus Xestospongia . Bioorganic and Medicinal Chemistry. 2010;18(16):6006–6011. doi: 10.1016/j.bmc.2010.06.066. [DOI] [PubMed] [Google Scholar]
- 9.Belarbi EH, Gómez AC, Chisti Y, Camacho FG, Grima EM. Producing drugs from marine sponges. Biotechnology Advances. 2003;21(7):585–598. doi: 10.1016/s0734-9750(03)00100-9. [DOI] [PubMed] [Google Scholar]
- 10.Schmitz FJ, Gunasekera SP, Yalamanchili G, Hossain MB, van der Helm D. Tedanolide: a potent cytotoxic macrolide from the Caribbean sponge Tedania ignis . Journal of the American Chemical Society. 1984;106(23):7251–7252. [Google Scholar]
- 11.Chevallier C, Bugni TS, Feng X, Harper MK, Orendt AM, Ireland CM. Tedanolide C: a potent new 18-membered-ring cytotoxic macrolide isolated from the Papua New Guinea marine sponge Ircinia sp. Journal of Organic Chemistry. 2006;71(6):2510–2513. doi: 10.1021/jo052285+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith AB, Lodise SA. Synthesis of tedanolide and 13-deoxytedanolide. Assembly of a common C(1)-C(11) subtarget. Organic Letters. 1999;1(8):1249–1252. doi: 10.1021/ol9909233. [DOI] [PubMed] [Google Scholar]
- 13.Jung ME, Marquez R. Efficient synthesis of the C1-C11 fragment of the tedanolides. The nonaldol aldol process in synthesis. Organic Letters. 2000;2(12):1669–1672. doi: 10.1021/ol005675l. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich G, Hassfeld J, Eggert U, Kalesse M. The total synthesis of (+)-tedanolide. Journal of the American Chemical Society. 2006;128(43):14038–14039. doi: 10.1021/ja0659572. [DOI] [PubMed] [Google Scholar]
- 15.Meragelman TL, Willis RH, Woldemichael GM, et al. Candidaspongiolides, distinctive analogues of tedanolide from sponges of the genus Candidaspongia . Journal of Natural Products. 2007;70(7):1133–1138. doi: 10.1021/np0700974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trisciuoglio D, Uranchimeg B, Cardellina JH, et al. Induction of apoptosis in human cancer cells by candidaspongiolide, a novel sponge polyketide. Journal of the National Cancer Institute. 2008;100(17):1233–1246. doi: 10.1093/jnci/djn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanif N, Tanaka J, Setiawan A, et al. Polybrominated diphenyl ethers from the Indonesian sponge Lamellodysidea herbacea . Journal of Natural Products. 2007;70(3):432–435. doi: 10.1021/np0605081. [DOI] [PubMed] [Google Scholar]
- 18.Aratake S, Trianto A, Hanif N, de Voogd NJ, Tanaka J. A new polyunsaturated brominated fatty acid from a Haliclona sponge. Marine Drugs. 2009;7(4):523–527. doi: 10.3390/md7040523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman CJ, Vegt PD, Debruyne FM, Vooijs GP, Ramaekers FC. Squamous and transitional elements in rat bladder carcinomas induced by N-butyl-N-4-hydroxybutyl-nitrosamine (BBN): a study of cytokeratin expression. American Journal of Pathology. 1985;120(3):419–426. [PMC free article] [PubMed] [Google Scholar]
- 20.Ulukaya E, Ozdikicioglu F, Oral AY, Demirci M. The MTT assay yields a relatively lower result of growth inhibition than the ATP assay depending on the chemotherapeutic drugs tested. Toxicology in Vitro. 2008;22(1):232–239. doi: 10.1016/j.tiv.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 21. http://www.brc.riken.jp/lab/cell.
- 22.Paul GK, Matsumori N, Konoki K, Murata M, Tachibana K. Chemical structures of amphidinols 5 and 6 isolated from marine dinoflagellate Amphidinium klebsii and their cholesterol-dependent membrane disruption. Journal of Marine Biotechnology. 1997;5(2-3):124–128. [Google Scholar]


