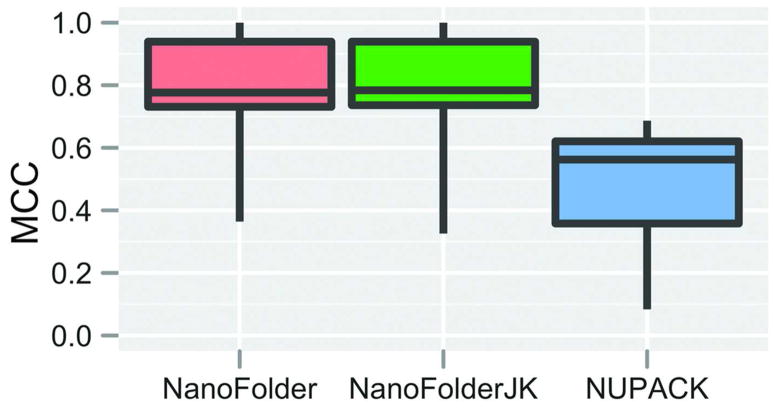
Figure 2.
Box-Whisker plot of achieved prediction qualities (Matthews correlation coefficients) for different methods applied to the set consisting of the nine multistrand RNA structures listed in Table 1. Each box-whisker element depicts the first quartile, median and third quartile in the form of a colored box; minimum and maximum scores are depicted as “whiskers” (vertical lines eminating from the colored box element). No data elements were considered “outliers”. The method NanoFolderJK corresponds to the leave-one-out (jack-knife) approach of training and testing the inter-strand interaction penalty parameter. For the method indicated as NanoFolder, this parameter was set to the median of the training results achieved in the jack-knife method.