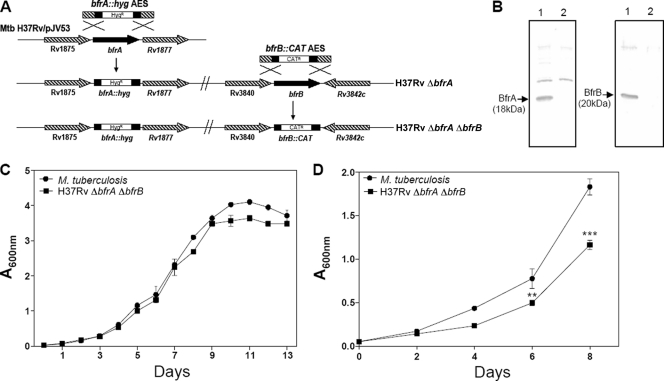
Fig 1.
Characterization of H37Rv ΔbfrA ΔbfrB. (A) Disruption of bfrA and bfrB genes of M. tuberculosis by using the recombineering method. The diagram represents homologous recombination between bfrA::hyg AES and the bfrA gene in M. tuberculosis to generate H37Rv ΔbfrA, followed by recombination between bfrB::CAT AES and the bfrB gene in H37Rv ΔbfrA to generate H37Rv ΔbfrA ΔbfrB. (B) Confirmation of disruption of bfrA and bfrB deletion in M. tuberculosis by immunoblotting. Five μg of cell-free protein extract of M. tuberculosis (lane 1) and H37Rv ΔbfrA ΔbfrB (lane 2) was loaded onto a 12% polyacrylamide gel and subjected to electrophoresis. BfrA and BfrB were detected by immunoblot analysis using anti-BfrA or anti-BfrB polyclonal antiserum. BfrA and BfrB proteins migrated as protein bands corresponding to a molecular mass of 18 and 20 kDa, respectively (lane 1). The disruption of bfrA and bfrB in H37Rv ΔbfrA ΔbfrB was confirmed by the lack of expression of either protein (lane 2). (C) Growth kinetics of M. tuberculosis and H37Rv ΔbfrA ΔbfrB in MB7H9 medium. Cultures were inoculated in duplicate with a starting absorbance (A600) of 0.025, and the growth was monitored for 13 days. (D) Growth curve of M. tuberculosis H37Rv and H37Rv ΔbfrA ΔbfrB in iron-depleted medium. M. tuberculosis H37Rv and H37Rv ΔbfrA ΔbfrB were grown in MB7H9 medium supplemented with 0.05% Tween 80, 10× ADC, and 100 μM 2′2′ dipyridyl. The growth of the strains was monitored by measuring the A600 for 8 days. There was a significant difference in the growth of H37Rv ΔbfrA ΔbfrB compared to that of M. tuberculosis H37Rv under iron-deprived conditions. The values of absorbance are represented as the means (± standard errors) of three independent samples, and the experiment was repeated three times. **, P < 0.01; ***, P < 0.001 (two-way ANOVA).