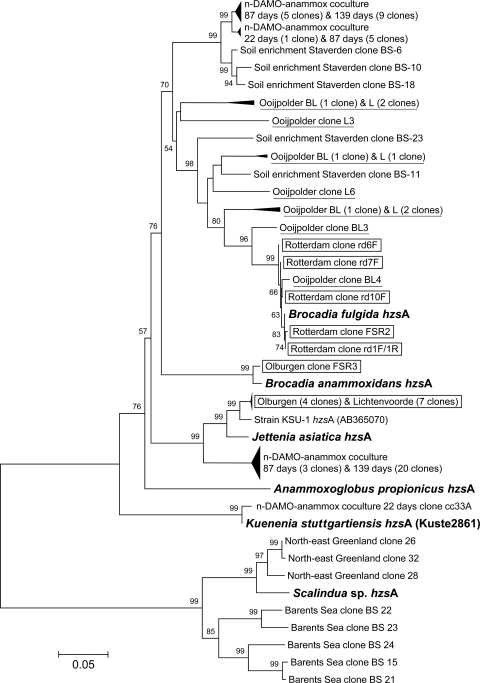
Fig 2.
Phylogenetic analysis of hzsA gene sequences from several anammox bacteria (in bold), reactor samples, and environmental samples (93 in total). Clones from full-scale Anammox wastewater treatment plants are shown in boxes, and clones from the agricultural land ditches are underlined. Bootstrap values of >50 (500 replicates) are shown at the branches. The neighbor-joining tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Jukes-Cantor method and are in the units of the number of base substitutions per site. The bar represents 5% sequence divergence. Codon positions included were first plus second plus third. All ambiguous positions were removed for each sequence pair. There were a total of 2,433 positions in the final data set.