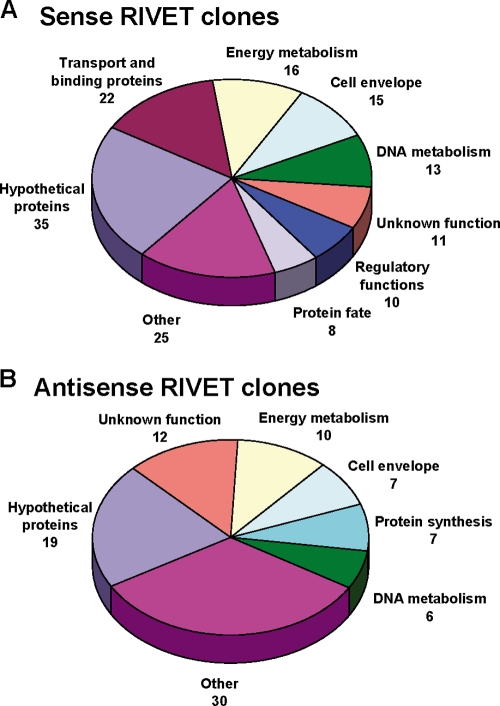
Fig 2.
Distribution of E. faecalis OG1RF sense (A) and antisense (B) RIVET clones isolated from a subdermal abscess infection. The E. faecalis genomic DNA sequence from each isolated RIVET clone was analyzed as described in Materials and Methods to determine the identity of the putative in vivo-activated promoter and the corresponding gene immediately downstream of the putative promoter. Each corresponding gene was assigned to a functional category by using the JCVI Comprehensive Microbial Resource (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi) for the E. faecalis V583 genome. All protein-encoding loci, as listed in Tables S2 and S3 in the supplemental material, were included. The number of genes in each functional category is given. The functional categories classified as “other” in panel A are as follows: cellular processes (5 genes); fatty acid and phospholipid metabolism (4 genes); pyrimidines, nucleosides, and nucleotides (4 genes); protein synthesis (4 genes); amino acid biosynthesis (2 genes); signal transduction (2 genes); transcription (2 genes); biosynthesis of cofactors, prosthetic groups, and carriers (1 gene); and mobile and extrachromosomal element functions (1 gene). The functional categories classified as “other” in panel B are as follows: biosynthesis of cofactors, prosthetic groups, and carriers (5 genes); pyrimidines, nucleosides, and nucleotides (5 genes); regulatory functions (5 genes); signal transduction (4 genes); cellular processes (3 genes); transport and binding proteins (3 genes); central intermediary metabolism (2 genes); fatty acid and phospholipid metabolism (1 gene); protein fate (1 gene); and transcription (1 gene).